As you all know, there have been two aDNA papers released recently about Central Asia to North India. I didn’t dedicate a post to them (there are comments in the previous thread about them, though), mostly because the first one (The formation of human populations in South and Central Asia, Narasimhan el al. 2019) had already been extensively commented when the preprint was out, and while it did bring more samples these mostly add quantity to already sampled populations with few new ones (and not relevant enough to deserve a new post), while the second one (An Ancient Harappan Genome Lacks Ancestry from Steppe Pastoralists or Iranian Farmers, Shinde et al. 2019) finally brought the first ancient sample from within modern India, but it was only one low quality one that didn’t add much to the better quality “Indus periphery” ones already present in the former paper.
However, there’s still a bit of confusion regarding the ancestry to the people of the Indus Valley (and generally to the genetic structure of SC Asian populations), so here I’ll try to give some insights that might help to clarify the situation for further, better informed, analysis.
The basic premise here would be to split Iranian ancestry into West and East Iranian. The main difference would be the ratio of Basal Eurasian to ANE ancestry (higher in the west, lower in the east), but given the lack of Mesolithic samples we’re still unable to get the whole picture. However, some basic concepts can still help us to better understand the situation. So let’s start.
Vahaduo’s online modelling tool
And I’ll use his post to introduce a recently released online tool that deserves more attention, given its quality and usefulness. It’s been written by Vahaduo, with a similar purpose to my own Xmix, but more complete, faster and not requiring any local installation. So I’ll use this post to show how to use it for any of the readers to be able to try their own models and be able to test for themselves whatever they are interested in.
The first (and only) thing you’ll need is to get some datasheets that are valid to use with Vahaduo’s program. The best (and recommended) ones being the Global 25 scaled datasheets from Eurogenes. One will have all the individual samples and the other the averages of each population. Ones you have these, you can proceed to the site and start testing. Here we’ll go directly to test what I mentioned above: East vs. West Iranian ancestry.
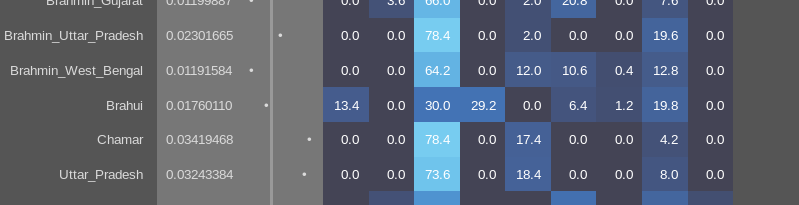
For West Iranian ancestry, I’ll use the average of the Early Neolithic samples from the Ganj Dareh site in the Zagros mountains. And for east Iranian I’ll use the average of the easternmost samples we have so far: Sarazm_Eneolithic. So I’ll need to copy the coordinates of these in the “SOURCE” tab (one per line):
Now, two sources will probably not be enough to test the samples from SC Asia and Indus periphery, since there are more streams of ancestry in them (at least one related to ANF and the other to AASI). So I’ll go ahead and add the average of Barcin_N samples and the average of modern Onge and Naxi populations.
Then for the targets, I’ll use individuals instead. In this case I’ll start with the “Indus periphery” samples, which are labelled in the datasheets as IRN_Shahr_I_Sokhta_BA2 and TKM_Gonur2_BA, so again one per line I copy and paste them in the “TARGET” tab:
And now we’re ready to run the program and get the results. Since we’ve added multiple target samples, we should go to the “MULTI” tab and click on the “RUN” button, which will show us this:
As you see, Naxi doesn’t appear in the results, and that’s because all the samples got 0% ancestry from it. If we wanted to see all the sources in the output, we’d just have to click on the “PRINT ZEROES -NO” button (which would change to “PRINT ZEROES – YES”) and click “RUN”. The “AGGREGATE – YES” button is to aggregate the percentage of multiple sources with the same label (for example if instead of using the average of Ganj_Dareh_N we would have used all the individuals as sources, we would choose to either see the results with each individual specified or to aggregate them into a single column with the sum of them).
Then we can download a .CSV file to import it into a spreadsheet and make further calculations if needed (or for sharing purposes using Google Docs, for example). The “DISTANCE” tab is also useful to calculate the distance between a sample to all the sources (you could copy for example the whole datasheet, being careful not to copy the first row with the PCA labels) and get the top 25 closest samples/populations.
It just takes some minutes to get familiar with the program and the options so go ahead and try it. It’s definitely a very useful tool.
Some insights into SC Asian and Indus Valley ancestry
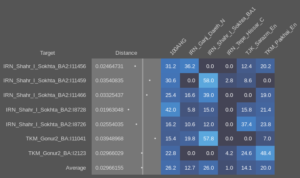
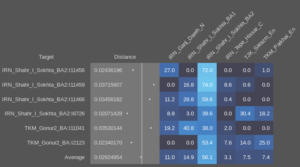
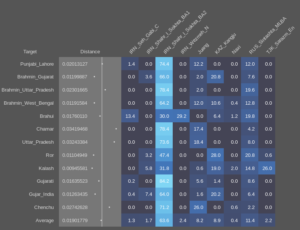
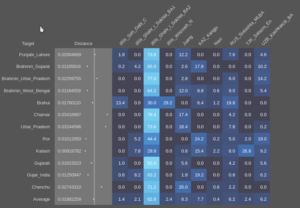
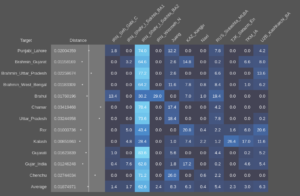
So let’s start with what we see in the above model of the Indus periphery samples. Leaving (for now) aside the fact that they may have some recent admixture from the places where they were found, one striking thing is the very variable ratios of West and East Iranian ancestry. In the following spreadsheet the above results can be seen (Sheet 1) together with a second run with the 100AHG simulation provided by Matt in the previous thread (Sheet 2), and in both the the calculated ratio of West to East Iranian ancestry. It’s easy to see that there is no correlation between that ratio and the amount of AASI in each sample, which makes it irrelevant for this matter whether they have any admixture from the local populations or not. Either way, we’re seeing a diverse population not just in terms of AASI to West Eurasian, but in the more sutle, but still important, West to East Iranian ancestry.
This pattern of significantly different ratios in West and East Iranian ancestry is equally seen in the regular Shahr-I-Sokhta BA samples (Sheet 3) and in the Turan Eneolithic samples (Sheet 4). The Iranian-like ancestry in the Indus periphery samples is therefor very similar to the one in those places. But they’re not all homogeneous, and point to mixed populations with probable input from West Iran. Modelling the samples with more proximate sources and using the 100AHG simulation again, it looks like this:
And using the sample with the highest amount of AASI as a source instead of the 100AHG simulation, something like this:
So what does this mean? First that things are a bit more complicated than getting the average of a population and building a tree estimating the divergence time from another one under an assumption that there is no admixture between them just because they’re not the same. In more simple words, we can’t really know with certainty if there was some migration from the Zagros Neolithic to North India or if there was none. Both options are possible. What we can say, though, is that we’re talking about a significantly different case to the Neolithic transition in Europe, since there must not have been a large replacement by outside farmers in any case.
All of this opens some interesting questions regarding the genetic history of South Asia. Unfortunately, we don’t have the data to give any answer to those questions, but it’s still worth knowing them and the different possible answers. For example:
- Who were the Mesolithic Hunter-Gatheres from North India?
- Who were the first farmers?
- Was there any subsequent migration before the Bronze Age?
Let’s break up the genetic structure of (putative) IVC samples into the 3 main streams of ancestry:
- AASI
- West Iranian
- East Iranian
This does not mean necessarily three different populations. Two or more of these ancestries could have been already mixed since very early. But let’s examine the possibilities:
First, a basic look at the geography of India tells us that there are no major barriers within it, compared to the big barriers with the outside. This makes less likely he possibility of two extremely different populations during the Mesolithic living in South and North India, and the one in north India being almost identical to the ones outside (Iran and Turan). It could be (only aDNA can tell us), but it looks like the least parsimonious.
Together with the diversity in the ratios of those 3 streams of ancestries, it’s really unlikely that we could be talking about an isolated India-specific population. We have to think in terms of some degree of migration to India from outside before the Bronze Age.
The possibilities about who was where at each point in time are many, and I won’t argue for any of them. It’s speculative at this point. But as possible examples:
We could have a AASI-rich population, but with significant East Iranian ancestry too during the Mesolithic. Then we could have a moderate migration from the Zagros Neolithic and no more migrations up to the IVC time where we have samples. This would be somehow similar to the Neolithic transition in Turan, where presumably a mostly East Iranian population was there in the Mesolithic and received some migration from West Iran during the Neolithic transition. The difference (apart from the lack of AASI ancestry in Turan), is that the communication between West Iran and Turan is easier, and gene flow continued (both ways) throughout the Chalcolithic and Bronce Age.
The problem with this scenario is how to explain the presumed differences in levels of AASI in the IVC and their lack of correlation with the East Iranian ancestry that would have been associated with it.
Scenarios were we separate the three streams of ancestry could better explain the situation, though given that Turan Chacolithic had already a diversity in East and West Iranian ancestry ratios that could serve as a single migration too (note that neither West Iran Chalcolithic or Turan Bronze Age would fit well as admixing sources due to their excess of ANF-related ancestry). I’ll leave to the comments any further variations within these constraints.
The Steppe ancestry in Turan and North India
This subject has already been discussed everywhere in great detail, for a very long time. So I didn’t plan to look at it again. I don’t have much more to say, but I’ll go through it fast.
The post BMAC samples that we have hardly show any steppe admixture. In the same spreadsheet linked above (Sheet 5), I’ve added the samples with an average date in calBP of <3700 years in descending order (note that the Present is defined as 1950 CE, so you’d need to add 69 years to get the real BP as of today). There’s one Parkhai_LBA_outlier (1497-1413 calBCE) that shows 9.2% Sintashta_MLBA admixture. The rest until the last BA samples (3250 BP) are in the noise levels. It’s only the single Iron Age sample from Turkmenistan (912-799 calBCE) that has a big increase to 50%.
In the Swat Valley, we have the earliest samples from the period 1200-800 BCE. They have significantly more steppe admixture, ranging between 20% and 0% and an average of around 10%. The variability of the amount of steppe ancestry doesn’t seem very compatible with their estimate of admixture happening 26 generations before in that same place, in that same population. But the shortcomings of their observations that provide evidence of the arrival of steppe ancestry to South Asia in the first half of the second mill. should have been already evident without looking at individual variability with up to 0% levels.
Another of the inferences for supporting such evidence was their observation that after he MLBA the steppe got Siberian/East Asian admixture, which is not found in modern India. However, they could model modern population using the Kangju samples from Kazakhstan (II-V CE). Modern samples are never a good way to make inferences about prehistory (including modern frequencies of certain uniparental markers). It seems rather arbitrary why would populations choose Sintashta or Kangju (though maybe Kshatriya ones make sense),
or indeed why would they choose Sintashta_MLBA or the Kashkarchi_BA samples from 1200-1000 BCE which are almost identical,
or even adding the Turkmenistan_IA sample mentioned above too as a source (as suggested in the comments from the previous thread), which further splits the steppe ancestry in a relatively random way.
Overall not much to add about all of this steppe part. We’ll have to wait to see those samples from the first half of the second mill. BC around the Punjab before we can know with certainty how all this went.



@Singh, I’ll be honest I don’t know what to think about that question – I’m pretty unsure about the process of estimating the early Neolithic / Mesolithic / late Upper Paleolithic Near Eastern populations as Dzudzuana+ANE/WHG/etc+extra Basal Eurasian vs just modelling those in other terms.
Basal Eurasian still needs consideration in light of whether it actually exists or there is something more like a trifurcation within Eurasia* and then it can be explained by low level back-migration of certain Near Eastern related groups to Africa, or from somewhere in Northern Africa to Africa past the Sahara and the Near East – https://imgur.com/a/v6h78bp
*Of course this is a simplification, as it minimizes deep splits in ENA that almost occur at same time depth as East+West Eurasian, Ust Ishim as close to trifurcation etc, but a simplifcation for the purposes of the Basal Qurasian question.
@AK, I’ve probably already commented re what I see as plausibility of late geneflow between South Asia and Central Asia, so I would only have to agree with that comment.
Narasimhan does tend to make the simplifying assumption that all groups in Central Asia had extensive East Asian related geneflow by IA, precluding them from any flow, but I’m not sure this is true or proportionate – https://imgur.com/a/Ie4ukyf – Xinjiang Tajiks today estimated to harbour very little East Asian ancestry, perhaps only 10% and Wusun and Kangju not necessarily outlier populations.
I still am left with the impression that Narasimhan’s paper does seem to be in a rush to try and find a Steppe_MLBA only source of ancestry for Swat and to preclude further Turan (with low Anatolian) ancestry to really push for a route via Central Asia without touching BMAC (for the reasons that this fits their particular linguistic story with no complications or exceptions).
But this does have obvious problems with what has been said before in the archaeology by Mallory and Parpola and others (with equal conviction to the supposed irrefutable archaeological links of Indo-Iranian to Sintashta and Andronovo) and probably some statistical weaknesses where populations with quite a bit of Turan CA->BA related admix probably can’t really seriously be excluded.
Even Narasimhan himself seems to in post paper comments be inventing some notional model where Indo-Aryan groups very rich in Steppe_MLBA entered South Asia via Swat but leapfrogged over any samples we can actually detect, then backflowed into Swat later in history in order to explain the patterns of decreasing relatedness in Swat to Turan over time and of R1a, without having to propose an alternative direction of migration which would entail going through other areas.
One thing I am pretty disappointed about now with Narasimhan’s paper is that the supplement promised that they would upload all their f stats onto the Data Visualizer
(https://public.tableau.com/profile/vagheesh#!/vizhome/TheFormationofHumanPopulationsinSouthandCentralAsia/AncientDNA) which seems to have fallen through. (Supplement refers – “The third and fourth tabs visualize f3- and f4-statistics respectively.”… but this wasn’t uploaded.)
Since patterns in those would really serve to test whether their models hold up, and that data is effectively closed, we’re reliant on hobbyists and other labs to run those stats and test the patterns in a broader sense (which the main prominent hobbyist at least seems not inclined to do, having moved away from publishing big sets of formal stats and towards proprietary PCA, being pretty happy with Narasimhan’s results and mentality and openness to other scenarios having probably hardened and closed).
@Matt
Yes, the actual modeling part of narsimhan’s paper is shoddy. He seems to have spent the least amount of time on this most important aspect.
Hypothesizing about a yet unsampled 50/50 steppe/IVC population as a source for modern indians while rejecting a post iron age inflow from the steppe is also insane.
I did tweet to him asking about the east asian affinity of Swat IA and modern NW indians. He brushed it off as Onge affinity, whereas Han is clearly selected over Onge as well as 100AHG.
which models (with some turan source) for SwatIA are you suggesting? ill test them out..
@Singh sry, cant help you there. havent studied the matter
@AK: I’ll have a look at Swat_IA and see what models I could think. I am thinking that trying to find stats driven by whether Anatolian came to Swat in association with CHG or WHG might help work out things to some degree (presumably coming with CHG would be expected to be more of a BMAC signature while WHG would be a European ancestry through Sintashta+Andronovo signal? Confound that CHG vs IranN probably distinguishes steppe ancestry).
On another tack just starting from the complete basics one thought if you’re looking at these things that may start you off on something interesting:
Quick exploration of the model fits for Indus Periphery provided by Narasimhan: https://imgur.com/a/adussq4
Two main successful models for In_Pe from Narasimhan 2019 – Model A: Ganj_Dareh+WSHG+AHG and Model B: Sarazm_En+Tepe_Anau_EN+AHG.
(These models use as outgroups only: Right South Asia: Ethiopia_4500BP.SG, WEHG, EEHG, Ganj_Dareh_N, Anatolia_N, WSHG, ESHG, Dai.DG)
We find that of the samples with higher coverage: I8726 works well with Model A and poorly with B, while I8728 and I11456 works well with B and poorly with A.
There is not a correlation visible of model fit with AHG level or the PCA position.
If you are interested in exploring them, and re-running Narasimhan’s models, you might see something obvious about why Model A works for I8726 but poorly for I8728 and I11456 and vice versa with Model B.
Like, does Tepe_Anau_En+Sarazm_En have too much Anatolian for I8276, or is it something quite different? E.g. not enough WSHG?
And likewise does Ganj_Dareh+WSHG lack Anatolian which I8728 and I11456 need or is it again something different?
One other thing that might be worth your doing and which Narasimhan doesnt is if you have the software to test the Indus_Periphery samples and Swat_IA in qpWave.
Basically qpWave will tell you how many “streams” of ancestry you need to form a set of samples (see – https://www.nature.com/articles/s41467-018-05649-9/tables/3 – for an example).
If they really do form a cline with respect to a set of outgroups, then you’ll get Rank=1 (which means 2 streams = simple cline), while if the Rank is any higher, they’ll be related by more streams than this, and Narasimhan 2019’s concepts that the Indus_Periphery form a cline, or the Swat_IA form a cline, may be unsound, and the basis for their cline extensions in doubt.
Lazaridis uses qpWave quite a bit in his papers.
“I did tweet to him asking about the east asian affinity of Swat IA and modern NW indians. He brushed it off as Onge affinity, whereas Han is clearly selected over Onge as well as 100AHG.”
This is something seen in the online tableau data from Narsimhan. Perhaps its reason they drew a tortuous path from steppe to India via Afnasievo and IAMC?
Are you going to make a post on the Italian paper, Alberto? Very interesting stuff there.
As I had suspected, Iron Age samples from Central Italy are largely similar (Etruscan and Italic). The difference is that most of the Latins have varying degrees of additional Bronze Age Anatolian or Armenian ancestry and thus cannot be modelled as a two-way admixture between Yamnaya and EEF. One sample from early Iron Age Latium as plots as ‘southern’ as Cypriots.
From the supplements:
In consideration of the inter-individual heterogeneity in this period in PCA and ADMIXTURE, we also performed admixture modeling for each sample separately. Based on the qpAdm results for all Iron Age samples collectively, we started by testing a two-way model with RMPR_CA and Russia_Yamnaya_Samara as the source populations and found that it provides reasonable fits (p>0.05) for eight of the 11 Iron Age individuals (Table S16) but can be rejected for R437, R850 and R475. We therefore tested for these three individuals alternative one-way, two-way and three-way models, if none of the simpler models fits. Based on one-way qpAdm modeling, R437 forms a clade with an individual from Croatia dated to the early Iron Age. In contrast, R850 forms a clade with an individual from Copper Age Anatolia. These two individuals both came from Latin archaeological context, together with four other samples, who can be modeled as two-way mixtures of Copper Age central Italian and Steppe-related ancestries.
Two two-way models fit well for R437 and R850: RMPR_CA + Armenia_LBA and RMPR_CA + Anatolia_IA.SG. In both models, the incoming source population is temporally proximate to the Iron Age Italian samples, and their geographic locations point to ancestry input from the Near East. Strikingly, R437 and R850 both carry more ancestry from the incoming source than the preceding local population, highlighting the substantial influence of this “eastern” influence on the genetic makeup of central Italians in Iron Age. Furthermore, the influence of this “eastern” ancestry is not limited to R437 and R850, as R1016 and R1015 can also be modeled as RMPR_CA + Anatolia_IA.SG, and R1016 (but not R1015) as RMPR_CA + Armenia_LBA.
—–
The Etruscan samples are just Yamnaya + EEF, with the exception of a woman who seems to have some type of African ancestry.
With the former it’s quite curious that one seemingly ‘Latin’ Necropolis had both a broadly Iberian-like individual (Yamnaya + EEF) and a broadly Cypriot-like individual (who derives most of his ancestry from Bronze Age Anatolia). What’s the explanation for this?
Marko, judging by Table S4, Anatolia.IA.SG is apparently the average of MA2197+MA2198, which they perhaps shouldn’t be using in their final models because the former looks ancient Balkan-like and the latter seens considerably mixed with something very ANE/ENA i.e. two very different individuals in the first place and neither likely representing the mainstream of Iron Age Anatolia. That their average works well might not tell us much here, other than that the average of those two might approximate the real ancestry involved due to it ending up mostly Balkan-like + Anatolian_MLBA-like (with a touch of something ENA)?
Their model with Armenia_LBA is interesting but I can’t help but wonder, with my cursory look, if R850 doesn’t represent something e.g. Aegean and they could only produce that model instead because it didn’t form a clade with the currently sampled Mycenaeans due to some perhaps not too significant differences in ancestry. On the PCA it seems to fall between Mycenaeans and Anatolia_MLBA, though I assume a model like that was already tested by the authors, so it will be interesting to test various models in Global25. Another issue is that Iron Age Anatolia itself might be already shifting towards the east compared to Anatolia_MLBA (compare contemporary Anatolian Greeks, so a similar effect as the averaged Anatolia_IA here but not exactly in the same way) but we’re lacking that kind of population in the current data. As such a hypothetical individual between Mycenaeans and IA Anatolia might be modelled better as something like IA Italy + LBA Armenia. I’ll draw a comparison to the also R437 which they can model either solely as EIA Croatia or CA Italy + LBA Armenia. Something similar could be going on here and influences from the Balkans and the Aegean sound a priori more plausible than something directly from Armenia and thereabouts for Italy, arguably.
Mostly trying to make sense of what this result could represent really though a straight up CA Italy + BA Armenia result would certainly be pretty interesting and the latter curious in what it might represent. The Y-DNA probably doesn’t help much since it looks like it could have been present in any area from Italy to Armenia? The mtDNA T2c1f does seem like a match to a BA Armenia sample but I have no clue how widespread it was/is, especially with the earlier waves of CHG-rich ancestry going west.
By the way, what’s up with the “Copper Age 3,500-2,300 BCE” Greece sample in Fig. S10? Do we have an unpublished individual from the period that’s something like 70% Anatolia_N – 30% Steppe_EBA in a future paper? Maybe it’s that Wang et al. “LN Greece” sample? It seems to comes out as something that might be intermediate between Croatia_IA I3313 and Bulgaria_IA I5769 in the K=5 supervised ADMIXTURE though, while the Wang et al. sample’s PCA position was Central European-like IIRC. Though counting samples in the “Bronze Age 2,300-900 BCE” Greece ADMIXTURE I see 14 individuals, the 10 Minoans + 4 Mycenaeans, which might indicate it’s just a misdating of Crete_Armenoi unfortunately rather than something from the most relevant period for Balkan IEzation.
A lot to take in for sure.
Sorry everyone for my long absence. I’ve been too busy and with not much to say, which made it difficult to keep up with the comments.
@Marko, Egg
I still didn’t read the paper about Italy. I’ll surely try to write a post about it, ut it won’t be immediately. Let’s see if the samples become available and make it into G25 so we can take a further look at them and add something to whatever the paper’s conclusions are.
For now, the fact that early Italics (not Romans) and Etruscans are roughly the same seems to make it difficult to draw some straight forward conclusions. We’ll need sampling from preceding era with good resolution to figure it out. But I’m hardly surprised that Etruscans are not any sort of recent migrants. It should be easy to say that Etruscan is a pre-IE language of Italy as a starting point and pick up from there.
Egg, I’ll comment more when I have the time to read the paper, but I’d agree that anything in Italy coming from Anatolia/Caucasus should be Balkans mediated. So better sampling of the Balkans is also going to be necessary to understand what was going on in Italy.
@Alberto
Would you exclude the maritime route directly from the Aegean coast or beyond? The Latin sample in question has way too much Iran-related ancestry to have come through the Balkans IMHO.
He probably just has some local ancestry.
I should be reading the paper much better first really but since we did discuss that one sample R850 a bit and I can’t help it, one other tentative idea I could throw out there in the interest of further discussion is that it might somehow represent a non-admixed Etruscan under the Aegean-Anatolian (vs the Central European) scenario from the area of the northwest Aegean, with the known remnant in Lemnos, and northeast Anatolia. I notice Frank suggests a possible scenario like that on Eurogenes.
In that case being able to model it solely as the Anatolia_Barcin_C I1584 in the paper could imply a northeast Aegean-northwest Anatolian origin (as Beekes relatively recently also argued) and the differences between Anatolia_Barcin_C and Anatolia_BA could be down to present BA structure rather than I1584 representing an outlier or an earlier situation that had changed in BA Anatolia, since all our BA samples and the other CA samples (Tepecik) are from much further southeast and so appear “southwest” on the PCA in comparison, in a bit of a cline. That in both cases we’re dealing with single individuals is unfortunate and makes me more skeptical about any particular scenario.
Marko
The effect of Cetina expansion would be interesting ; but we’d need samples from eastern Italy
I really don’t know if R850 could have come straight from Anatolia. Maybe. But people arriving by boat would be fewer than those arriving by land, I would guess?
In any case, that’s a Latin sample, not an Etruscan.
Alberto, it does come from an apparently Latin Necropolis but the question naturally still remains in what it represents when it plots between Mycenaeans and Anatolia_MLBA on their PCA, unlike the main Iron Age cluster. I’m not really sure why they didn’t also test models involving the main Iron Age cluster but only ones with CA Italians, just to cover all bases, but I suppose their scenario is essentially the same. It does look like you might be able to explain it as IA Italy, or even more specifically the fellow Ardean R851, + BA Anatolia based on position at least. With steppe-less CA Italians, the more northeastern Armenia_LBA (or the unfortunately averaged Anatolia_IA) will be naturally preferred.
From the rumours and leaks, I’m expecting a second paper from Central Italy with more information about all of these things that are not entirely clear in this one. Usually the papers dealing with the same subject from two different teams come out almost at the same time, so I’ll wait and see if that second paper comes out to make a post about both with more complete information.
In the meantime I’ll comment here anything I find interesting about this published paper, which I still couldn’t read in detail.
After looking at the available samples, and given that no further ones have been published yet, a brief comment about the IA ones:
I’ve been unable to find any significant difference between the non-outlier Etruscan and Latin samples. The number is too low anyway. But with these and with the data we have from unpublished ones the pattern should stand.
The preceding Bronze Age is still not clear, but for what we know and what we’ve heard, we can say that the R1b-L51 people didn’t have such a dramatic impact in the Italic peninsula as it did in Western Europe. Nevertheless, it’s probably not way too far either. The steppe admixture and prevalence of R1b-L51 is still very significant. Probably not enough to think that they replaced all or most of the Neolithic languages, but neither insignificant enough to say that they brought no language at all with them. There probably was a diverse linguistic landscape by the late BA, with both Neolithic and BA (steppe-related) languages existing in different areas (though we’d need good sampling to have a more precise idea).
The introduction of Italic languages was also not very dramatic, but obviously quite significant. Maybe around half of the people shifted to Italic and the other half didn’t (very roughly speaking). The thing is that from those who didn’t none of them spoke an IE language.
This is consistent with the data from Western Europe, where we have a large part of the population (in the areas where we do have attested languages) speaking non-IE languages after the Celtic expansion.
At this point, the idea that the steppe/R1b-L51 spoke an IE language is left without any supporting data. One would have to argue that this large migration brought a ghost language to Western Europe and Italy that we have absolutely no evidence of, but that it actually was IE (because it was). It just vanished without traces, even though many Neolithic languages stayed alive and well.
Nor does the case of substrates help in any way. First because it’s something quite difficult to deal with (I elaborated about it already), but even if we trust this difficult data the evidence is lacking.
To mention a different area from Iberia, the strong substrate is Insular Celtic has long been known. The theory was, if I’m no mistaken, that Celtic migrations had a lower impact in the British isles than in the continent, allowing for a stronger substrate. I don’t know if it was lower than in France, but it certainly was low. Now, this substrate has long been though to be Semitic (or some sort of Afro-Asiatic), with a second option frequently cited being a Vasconic language (the bias in the amount of data to work with when it comes to Afro-Asiatic languages vs. Basque is so big that it’s not surprising that it’s been easier to find correspondences in AA than in Basque). For example, see “The substratum in Insular Celtic“, R. Matasović 2012. Now that we know the population history of the British Islands it becomes rather impossible to argue that the pre-Celtic population spoke an Afro-Asiatic language, and equally difficult to argue that such significant substrate goes back to the Neolitic farmers.
More when we get the rest of the samples (or more data from West and/or SC Asia, whatever comes first).
Alberto,
Could you elaborate and explain your argumentation in more detail that led to your conclusion below based on IA Italy aDNA, as I’ve crucially not been able to follow your reasoning in this to completion, despite reading your latest comment several times.
“At this point, the idea that the steppe/R1b-L51 spoke an IE language is left without any supporting data. One would have to argue that this large migration brought a ghost language to Western Europe and Italy that we have absolutely no evidence of, but that it actually was IE (because it was). It just vanished without traces, even though many Neolithic languages stayed alive and well.”
Secondly, regarding your statements on Insular Celtic, are you saying that because the Neolithic in the British Isles was practically replaced genetically, there would have been no linguistic continuity of it detectable (in the present), and that the large non-IE substrate in Insular Celtic would therefore be associated with the largest post-Neolithic introduction, presumably steppe admixture? Or have I misunderstood entirely?
If not the steppe, what then could have introduced Celtic to the Isles? Or do you not expect it to be detectable as admixture?
Likewise, what could have introduced Italic to its present geography? When approximately? Do you think this would be discernible in genetic terms?
And do you expect some shared genetic ancestry between Celtic and Italic speakers, whether because you think Italic-Celtic would have once existed in joint form or otherwise is a valid language subgroup within IE, or even merely on account of shared roots in PIE?
And if you do expect shared genetic ancestry between the two groups of speakers, where in geographical terms do you now think this may have been mediated from, if not from the steppe? I’m not asking about your current supposition regarding the homeland, because that may not have become apparent to you merely from tentatively ruling out steppe/R1b-L51 based on acquiring the additional information from the (limited) IA Italy samples we now have. However, do you think that the aDNA we currently have points to (or at least doesn’t rule out in like manner) any potential intermediate stopover places indicated by the genetic ancestry of ancient or even current Italic, Celtic and possibly other IE speakers? Or do you not expect any common genetic ancestry to be all that clearly apparent due to a long and different series of admixtures, at different ratios depending on circumstances, for each ancient group of lE speakers, since spreading out from a linguistic homeland?
@ak2014b
I believe Reich et al. are working on a paper that documents rather significant genetic change in south-eastern Britain around the time one would expect Celtic languages to have arrived in Britain under the Hallstatt model.
https://www.bbc.com/news/science-environment-43712587
Should be interesting.
@ak2014b
Yes, I think you basically understood correctly, but to elaborate a bit more to clarify:
What these samples and other unpublished ones that we have already information about for some time show is that there is no genetic difference between Italic speakers and non-Italic ones (or more specifically Etruscan ones). They are both a similar mix of R1b-L51-steppe derived populations and Neolithic farmers, and predominantly R1b. Basically like in the rest of Western Europe (albeit with higher diversity and outliers).
Italic and Celtic languages must have formed (in a pre-proto-stage, some sort of Proto-Italo-Celtic sprachbund) around the North West Balkans/North East Italy to Eastern Alpine region in the MLBA. Each of the two proto languages (Proto-Celtic, Proto-Italic) being being from relatively nearby regions around the final BA. There is no relationship between these two languages (their genesis and expansions) and the much earlier (LN/EBA) migration of Bell Beakers to Western Europe and Italy.
When these languages expanded in the early Iron Age they replaced many of the preceding languages of Western Europe and Italy. But not all of them. We have a large part of the population still speaking other languages, but all of them are non-IE (with rare and unclear exceptions like Lusitanian).
So the question here would be: if Bell Beakers brought by far and large IE languages to Western Europe and Italy, where are those languages? How is it possible that we have absolutely no evidence, direct or indirect about them? The answer that they were all replaced by Italic and Celtic doesn’t solve the problem, because you’d need a sort of selective replacements of those languages, something statistically close to impossible. So on what base can we argue that Bell Beakers spoke an IE language? Why not Altaic? Or any other language family of choice?
Regarding the substrate in the British Islands, yes, the rationale would be that Bell Beakers had a very fast and large expansion there leaving very, very little chance for language survival. Much less after almost 2000 years, when Celtic arrived. So how could on explain a strong non-IE substrate in Insular Celtic if Bell Beakers already brought an IE language with them? Again resorting to almost impossible language survival of pre-BB populations and disappearance of BB languages? any argument becomes just too complicated and purely speculative. The evidence is that there is no evidence for BB being IE speakers, and it’s difficult to argue with no evidence against evidence.
Re: the ancestry of proto-Celtic and proto-Italic speakers, it’s a more speculative question and not important in the above context since their languages spread without a big genetic impact on the general population (though with good resolution and specific sampling we might get to know about it). My own idea is that they were largely from the Balkans (more directly from the NW Balkans, but in turn from the East Balkans), and therefor they should show some amount of West Asian admixture. The upcoming study mentioned by Marko above (@Marko, thanks! Very interesting, let’s see what the results of it show when it comes out) may give us some clues, though we’ll need to trace that ancestry in space and time until it got to SE Britain.
@Alberto
Thank you very much for explaining. I believe I finally understand.
Very good points, much to mull over. Every time I think (L)PIE has been resolved, you or some other person here continue to bring up other ways of looking at the same data that lead to different conclusions.
> there is no genetic difference between Italic speakers and non-Italic ones (or more specifically Etruscan ones). They are both a similar mix of R1b-L51-steppe derived populations and Neolithic farmers, and predominantly R1b.
You bring up R1b. The current study didn’t have an R1b Etruscan. On the other hand, the study only had 3 or so samples from putative Etruscan contexts (depending on if the Villanovan was counted or not). So you’re referring to the mentions of unpublished R1b in Etruscan samples, I think?
@Marko
Looks very interesting, indeed, thanks. With 1,000 samples no less. There’s no indication on when the study will be published, though. But the news report is dated April 2018, so hopefully the wait won’t be too long from here on.
Alberto: The answer that they were all replaced by Italic and Celtic doesn’t solve the problem, because you’d need a sort of selective replacements of those languages, something statistically close to impossible.
Eh, statistically speaking, it’s not like you’ve got tens or hundreds attested data points and they’re all independently distributed. It’s like 2 languages (Iberian and Aquitanian->Basque), both from a pretty similar geographic range, and only 1 of these was actually anciently attested and only 1 of these attested post-Rome. If we were to take as a random example that there were 4-6 languages, 2-3 IE, 2-3 not, some geographic buffer or such why the 2 above were protected, it’s not really very outside of probability.
For most “Celtic” and “Italic” languages anyway, there just isn’t enough data to actually say to what degree they are well defined by being called “Celtic” or “Italic” anyway, rather than something else that branched upstream (or is just not well described as being linked to these by tree like branching). There isn’t really enough to do either reconstruction based on extended lexicon, or on characteristic changes in phonology and grammar. Going back again to Garrett’s paper on apparently proto-Celtic innovations that are not present in certain continental Celtic dialects when attested, and same in apparently proto-Italic innovations which are not present in the limited evidence of some of the early branching “Italic” languages like Venetic (and implying must have spread through non-tree like convergent processes, like contact or in some cases homoplasy motivated by deeper factors). (“What is crucial in this model is that at some early date – say, at the beginning of the second millennium BCE – the dialects that were to become Celtic, or Italic, or Greek, shared no properties that distinguished them uniquely from the other dialects. The point is not simply that innovations could spread from one Indo— European branch to another: this is well known. The point is that while there was linguistic differentiation, the differentiation among dialects that were to become Celtic, for example, was no more or less than between any pair of dialects. At this time, there was no such thing as Celtic or Italic or Greek. “)
@Matt
It’s all the Mediterranean Iberia that was non-IE, from South Portugal to Catalonia. Unless you consider Tartessian an IE language. And then Aquitaine too, in Atlantic France. But here I’m adding the Italic Peninsula too, where several non-IE languages are known from the Iron Age.
So my question is: if all of these non-IE languages cam from Neolithic farmers, where are the IE languages of the Bell Beakers? One can argue that they disappeared by chance so we don’t know about them, but at that point how is that compatible with arguing that they were IE?
About Garret’s paper we’ve discussed before. I can’t see his Celtic example as something realistic. And I don’t think I’m the only one. IIRC, when I mentioned that among other things there would be no explanation for the border between Celtic and Germanic you answered something about Roman empire (?) to explain it that I didn’t understand, since Romans had not even moved from Italy at that point. But even when they did, it seems irrelevant for the problem. But that’s just one of the many reasons why such scenario is untenable and why no one has cared about it. It’s just a theoretical exercise without any realistic evidence to support it.
@Alberto, I’d presume that IE languages of the Bell Beakers would probably be at the very least Celtic and Italic, even if they are presumed to then expand much later. Divergences from other IE (inc Germanic) by Italic and Celtic must happen by around 2500 BCE in any model after all. (I know that there are various complicated arguments that Celtic-Italic *can’t* be ultimately from the Bell Beakers but must be somehow from Southeast/Northeast Europe somehow, but it seems a bit of a stretch).
I can’t remember what you said about Celtic and Germanic so I can’t comment too much on that? I would say that the existance of non-convergence areas where there were actually boundaries does not refute that convergence areas existed.
What exactly do you reckon is untenable though? Linguistic sprachbunds and convergence areas in general? They’re pretty obviously not. Specifically that the languages we call Celtic today are descended from a wider set of IE languages in Western Europe that were part of a convergence area? We see all the time collections of present day dialects which *all* share an innovation which was not present in their “common ancestor” and which spread unevenly through them. Nothing unrealistic about it whatsoever (if would be hard to think it was unless one had literally never heard of linguistic convergence!). I think tons of folk are interested in when and where the Celtic language linguistic innovations actually arose and spread.
@Matt
I’d presume that IE languages of the Bell Beakers would probably be at the very least Celtic and Italic, even if they are presumed to then expand much later. Divergences from other IE (inc Germanic) by Italic and Celtic must happen by around 2500 BCE in any model after all.
But why would you presume that those languages (or better to say the ancestor of those languages) were from the Bell Beakers that lived 1500 years earlier? I don’t see anything in the languages or in the archaeology that links them specifically to the Bell Beakers. Why couldn’t they be from the Corded Ware Culture, or Unetice, or some BA culture from Hungary (or from anywhere else, really)? That seems like a random pick to me.
But ultimately it doesn’t even solve the problem of which languages were brought by the Bell Beakers to Western Europe and Italy. Were is the evidence to support that they were IE, an evidence strong enough to outweigh the evidence for them being non-IE? We don’t want this to turn into a new R1b from the east or from the west kind of debate, do we?
What’s untenable is Garret’s explanation of Celtic (but not only Celtic, he mostly refers to all except Romance sub family) that they formed in situ by language convergence from an old form of PIE. That this language undifferentiated from the other ones that would become Italic, Hellenic or Balto-Slavic developed after its arrival to Iberia, France and British Islands (mainly, but not only) by convergence throughout these areas and divergence with the neighbouring ones.
I’m a great supporter of language convergence, and have dedicated a whole post to it (mostly) a while back. Including convergence in Romance languages that had long been neglected by linguists. But that doesn’t mean that I could ever argue that Spanish, Italian and Romanian (among others) belong to the same family because of convergence without any proto-language ever existing and expanding. Not only because we have obvious evidence that they don’t (in the form of the Roman empire), but even without such well known evidence it would be untenable I think that for clear enough reasons to not need to elaborate.
@ Alberto
Yes, we need Bronze Age samples, from both the BB derived North/ west & Balkan-influenced Eastern coastline.
W.r.t. Garrett / Chang, i think it is a useful concept, and that’s all it is. E.g., we can propose that some languages formed in situ, e.g. Italic formed in Italy due to 3-4 streams of influence, with one overriding, & others forming substrata, etc
To hard data; we cannot escape the long noted observations that Primitive Irish is closely similar to pre-Roman Age Gallic. One doesn’t need any ‘stats’ to dispel the myth that it arrived with BBC (which in any case is a fringe theory).
This will certainly nicely dovetail with the archaeological evidence & genomic data Marko just mentioned
On the other hand; it could always be the case that it just so happened that BB switched languages in southwest Europe (the Midi; Liguria; Iberia) and those few in the Netherlands retained their real language (which alas was not recorded for 3000 years later)
@Alberto, re comparison with Romance languages, the points of comparison are though:
– That the subgroupings of Romance languages today do not exist because each shared a separate ancestor after Latin, but because different groups of initially undifferentiated Latin dialects shared in regional convergence after divergence from Latin. That is French, Spanish, etc. exist as separate languages because of convergence, not because each had a separate ancestor “between” Latin and the present day language.
And note that if we talk of Celtic, Italic, etc. if you adopt the steppe timing standard view, these are at the time of Italic (Latin) expansion, languages which should have a time depth of differentiation on the order of what French, Spanish, have today (and would have only had 1000 years of differentiation by the Late Bronze Age, suggesting not even mutually unintelligble at this time). So this is not an unreasonable analogy. (It’s a better analogy than to suggest that IE languages in Western Europe back in 1000-0 BCE had 4000 years of differentiation behind them, as say French-Farsi today! The only way the analogy is bad is if we reject a steppe date for an early neolithic one.).
– Beyond convergence effects which have defined *separate* Romance branches, if you tried to reconstruct of *all* the common ancestor of Romance languages based only on modern evidence, you’d probably reconstruct a “proto-Romance” which was wrong, and crucially much further from proto-Indo-European, than the real Latin was, with lots of “proto-Romance” features which actually spread later due to contact and convergence.
Even if we knew of Latin, if we didn’t know the detail of how features actually spread by convergence (within Romance as a whole and subbranches), we might imagine a set of structured waves of “proto-Romance people” that “replaced” Latin speakers to explain all the features we see in the language. (Seems pretty laughable that anyone would think this, but they probably would propose it, if we didn’t actually know how it happened.)
In the same way, if you used attested Celtic languages at the time of try and estimate an ancestor, we may be thinking it included many common innovations which spread later, to the extreme that the actual language that was the genetic ancestor may have had no specifically Celtic innovations on LPIE, and that *all* these spread through convergence.
Matt, yes, if we tried to reconstruct Proto-Romance from modern languages we’d end up with something quite different from Latin, especially in the grammar. But that obviously doesn’t mean that some Romance population replaced the other branches of Latin descendants. That would be a wrong conclusion. Just as wrong as it would be to argue that a Proto-Romance lanuage (Latin) never existed or expanded, and that Romance languages acquired *all* of their specific features and innovations from an archaic form of LPIE through language convergence. The latter is indeed specially laughable.
Just as it is to argue that Celtic languages formed in parallel (i.e, simultaneously) from a LPIE dialect that arrived c. 2500 BC in Iberia, Great Britain, France and North Italy through language convergence between themselves and divergence from other neighbouring languages that formed other families.
But all of this seems irrelevant to the main question and I’m not really sure anymore what are we arguing about.
@Alberto: Just as wrong as it would be to argue that a Proto-Romance lanuage (Latin) never existed or expanded, and that Romance languages acquired *all* of their specific features and innovations from an archaic form of LPIE through language convergence. The latter is indeed specially laughable.
Let’s think about why that is “laughable” though. Because we have extensive primarily linguistic evidence that Rome did expand, and we have evidence that there are all sorts of IE dialects about that would refute the features involved being a product of convergence over that area, and we can add to that that would be talking about a time depth of 4-5 thousand years, not around 1-2kya.
It’s not “laughable” that linguistic convergence could operate, inherently, simply the case that in that example, we have available which is directly contradictory. We know it’s wrong because to see Romance as purely the product of convergence effects in PIE dialects since dispersal *because* we have evidence that they’re not, not because the idea is wrong. Though the longer the timescale, the harder to imagine it’s true – a convergence effect creating a variety over 1-2 kya is quite easy to think of (and even obviously occurs within Romance to define subgroups and the family as whole today, as per upthread) but over 4 kya it is a push to imagine that chance isolation and divisions wouldn’t break it apart.
That’s why people who are eminent within the field of Celtic linguistics like Koch can find ideas of the formation in situ during 2500-1000 BCE from undifferentiated (or minimally differentiated) LPIE, plausible rather than simply refuted (as in Koch+Cunliffe’s “Celtic From the West” 2016 edition – https://i.imgur.com/OEZw7GB.png), and why this is one of the examples that linguists like Kalyan who look at convergence are interested in (https://pdfs.semanticscholar.org/84a0/334086b1a1dfa75fc147887fbd1c55375b1c.pdf).
But yeah, this is all a bit of a tangent; the main point I wanted to make was that it actually doesn’t seem to me to be statistically too improbable that you would have Copper Age IE dispersal in Western Europe and then no attestation of clearly non-Celtic / non-Italic languages by the late Classical Era (when writing emerges), because the record is so poor (writing is late, limited), and the spread of Romance (then Germanic, etc.) so profound. Our ability to classify most of the IE variants which were even know about through classical records which survive (let alone what would have been lost) in terms of position on an IE tree is very limited, by the limited corpus.
@Matt
the main point I wanted to make was that it actually doesn’t seem to me to be statistically too improbable that you would have Copper Age IE dispersal in Western Europe and then no attestation of clearly non-Celtic / non-Italic languages by the late Classical Era (when writing emerges), because the record is so poor (writing is late, limited), and the spread of Romance (then Germanic, etc.) so profound.
Yes, this is the basic point, that there is no attestation of clearly non-Celtic / non-Italic languages. But poor as our record may be, there is attestation of a good number of non-IE languages. This is the evidence that we have.
I think we should leave it there and let each one decide for themselves what to think about it.
There must still have been some switching & survival in SWE; because those various languages aren’t really related genetically
@Rob
Yes, that’s almost definitely true for Italy (even if the poor knowledge of those non-IE languages may not allow us for too conclusive assertions). But at least the genetic data there would also support a significantly higher probability of survival of languages from Neolithic farmers.
In SW Europe, while the data we have suggests that it’s more likely that Basque and Iberian are related than not related, we still have Tartessian to deal with. The current knowledge does not allow us to establish any relationship to Iberian, but in an older post I said that (speculatively) I preferred to think it may be related for the advantage that this brings to the possibility of Bell Beakers still being IE (a door I didn’t want to close). Though now with the data from Italy it seems less relevant since we already have a consistent pattern of all being non-IE without genetic differences in their speakers vs. IE ones.
Rather ironically, I see steppists arguing against a relationship between Basque and Iberian, without realizing that the more unrelated non-IE languages we find, the lower the chances for BBs to have been originally IE. Not that it matters in the whole scheme of things, but still perplexing.
Iberians and now even Etruscans have a significant steppe/bell beaker component. Its consistent with a Scenario of a late bloom/spread of IE in Europe that’s not significantly correlated with a genetic component. Just like ADNA samples represent many ancestors, Inscriptions and literacy cannot happen in vacuum and is indicative of a large illiterate speaker base.
Some bell beakers may have spoken IE but they may not have been a majority.
“Beakers brought a Basque+Iberian language” requires a really specific set of relatedness claims.
The language would have to be young enough to have got there about 2000 years before it was attested, yet also gone through huge and extensive changes that mean that this has eluded virtually all specialists through the 20th century.
Despite languages still being fairly close together in space and sharing contact phonological features (which is fairly agreed upon I think). It has to be young as a deep relatedness of a vintage more like Uralic-IE is obviously not consistent with “Beaker dispersal” and in fact would refute it (a small group of Beakers obviously could not have brought different languages which were a clade with each other dating to thousands of years before).
At the same time, the language that is supposed to be associated with the huge and fairly hegemonic Beaker culture, and from which they would not switch, must have also been not attested anywhere else more widely within Europe by the time we get to writing. This requires a probably more extraordinary claim of replacement than that some Beaker descended cultures would have switched at the edges of their range, and early introduced IE languages being levelled out by subsequent changes…. And that would require a more clearly asserted mechanism of replacement (maybe stronger than “Probably elite recruitment and small migrations, somehow”).
…
Slightly off topic, but linked to the linguistic topic and how numerals have come up, I was having a look at a paper by Mark Pagel in 2013 – “Ultraconserved words point to deep language ancestry across Eurasia”. That his this to say about numerals: “The numeral words, despite having some of the slowest rates of lexical replacement in the Indo-European languages, have cognate class sizes of only two and do not appear in Table 1. Our conservative coding might have contributed to this, but number words are known to change among language families. These words can be invented independently, or because of their importance to communication and administration, they might be replaced en bloc and possibly at times of political or social unrest, as has been true historically of words for months of the year.”.
So numerals show these unusual patterns where in general they change very slowly, but can also be imported or changed all at once (as in the MSEA linguistic area!) – supportive of being compatible with either a very old relationship, or a sudden importation. Classes of the lexicon which tend to be conserved over 2000 year time scales, but which are not “ultraconserved” might tend to pose a better test for recent relatedness in that time frame.
Another more off topic aspect of Pagel 2013 that’s interesting is that by using “ultra-conserved” words and assuming a relationship, they attempt to date divergence of language families. They come to divergences from IE of (years): Dravidian – 14500, Kartvelian – 13000, Altaic & Chukchi-Kamchatkan & Inuit-Yupik – 12150, Uralic – 11700.
However, that’s based on Grey+Atkinson’s date of PIE splitting 8740 YBP and LPIE splitting 7300 YBP. If you assume that PIE split at 6500 YBP and LPIE at 5330 YBP (as Chang does, or more generally a general Copper Age steppe date), then divergences from IE of (years): Dravidian – 10800, Kartvelian – 9600, Altaic & Chukchi-Kamchatkan & Inuit-Yupik – 9000, Uralic – 8680. Effectively, mesolithic split dates for most (Uralic-IE fairly late mesolithic).
(Expressed as multiples of the intra-IE split, split from IE: Dravidian 1.7x, Kartvelian 1.5x, Altaic & Chukchi-Kamchatkan & Inuit-Yupik – 1.4x, Uralic – 1.3x. Or as LPIE: Dravidian 2x, Kartvelian – 1.8x, Altaic & Chukchi-Kamchatkan & Inuit-Yupik – 1.7, Uralic – 1.6. E.g. Dravidian is *only* twice as different from an extant IE language as that language is from another IE language, barring regional convergence effects.)
Btw, you guys will be interested in this, though it is small scale – https://www.biorxiv.org/content/10.1101/849422v1 – Gene-flow from steppe individuals into Cucuteni-Trypillia associated populations indicates long-standing contacts and gradual admixture
That’s a neat little study.
Would need some male samples to further evaluate dynamics (eg introgresion of I2 and R1 into late Tripolje society).
@ Matt
Iberian could be a pre-Beaker language, let’s say. E.g. being at the very extreme of their range expansion, and El Argar (its immediate successor in SE Iberia) was indeed heriarchical and almost ‘Apocalypse now’ scenario. But what about Acquitaine ?
The later Ceticization of Europe isn’t too odd . The various post-Beaker expansion cycles are very well recognized; and these account very neatly for the language strata in SW Europe
@Matt (don’t say i didn’t warned that we should probably leave it there – now you’ll have to read a long and maybe not too valuable reply, since we’re probably just going around in circles).
“Beakers brought a Basque+Iberian language” requires a really specific set of relatedness claims.
I agree. That’s why we can’t say with any certainty that Basque and Iberian were brought by Bell Beakers. The problem with finding their relatedness is, on the other hand, absolutely expected given the two languages we’re talking about. I’ve written in an earlier post about it, so I won’t reiterate the points made there. Maybe it’s easier to understand if one thinks of how difficult it would be to reconstruct proto-Germanic if the only known Germanic language was modern English, and how to show that it was related to other languages spoken in North-Central Europe if all we had from them were a few inscriptions with mostly personal names. Even this is not a fair comparison, since modern English is still a better proxy for Proto-Germanic than modern Basque for a putative Proto-Ibero-Vasconic. Think that while we would be talking of a language that spread 4500 y.a., Iberian scripts are from 2000 years later, and would still be relatively close to the Aquitanian spoken at that time. But modern Basque is another 2500 years later than that and (if we actually knew the Aquitanian language from 500 B.C.) mutually unintelligible with Aquitanian itself. So this is trying to reconstruct from modern Basque a language that predates it by 4500 years, with the only possible help of those few inscriptions in Iberian.
And just think about the Ogham inscriptions found in Scotland. They’re 99% chances in a Celtic language (very) closely related to Old Irish, and yet we can’t understand them.
It’s not like we’re talking about two well known modern languages (like those from SE Asia and China) were we know that numerals are a borrowing because we have the whole languages to compare them. And think how many languages are related to others, have contacts and borrowings from other languages, etc… We’ talking about thousands of cases. And how many borrowed the whole numeral system in spite of borrowing other words or even some number? Less than 1%?
And similarities are not limited to the numbers as you know. Phonological and morphological similarities are found too (the latter is almost a miracle given the knowledge we have about Iberian).
By the end of the day, this is a question that we cannot answer with certainty because we don’t have enough data. But given the latest research, it’s become rather difficult to argue for a non-relatedness even among the specialists, who (those who have tried) have been unable to argue against it in any convincing way (and when the main researcher who has been making progress in finding the similarities comes from a background of considering Vasco-Iberismo basically a pseudo-science, just to have to change his position in favour of it being a probable reality after years of research). Now that we have genetics telling us that it’s very easy to imagine that the languages are related, since the people who spoke them were closely related too, it becomes even more difficult to argue that it’s more likely that they are unrelated. It clearly isn’t. By far, it’s more likely that they are actually closely related. But we can’t say it for sure.
Anyhow, it’s an irrelevant question when it comes to anything IE or whether Bell Beakers spoke IE or not. For this my main point above is that we have good evidence of Bell Beakers being non-IE, based on the acutal languages found in former Bell Beaker areas, while the evidence for them being IE is completely absent. For me that’s what really matters, beyond any speculation or imaginative way to work around this evidence.
By the way, yes, it’s an interesting paper about the Cucuteni-Trypillian Culture. We’re missing the supplements to get a more precise idea about the samples, and being 4 females is not too much for today’s standards.
I hope we soon get a good sampling from Ukraine from the 5th and 4th mill. The 5th should see the advance of CTC farmers onto the steppe, I guess. The Alexandria (East Ukraine) sample from ca. 4000 BC has some 30% admixture from them, and he doesn’t look like an outlier given what we have from later periods. And then the arrival of the Progress-Yamnaya type of people from the east. Many details to sort out still regarding what exactly was happening there.
@Alberto, though we’re not talking about linguistic reconstruction though (easier with many daughters, and ones that aren’t interacting, but, always hard) rather simply systematic comparison of grammar and lexicon demonstrating genetic relatedness!
*Reconstructing* proto-Germanic from English and German alone means reconstructing the sound changes that led to the present form from a shared ancestor probably impossible, and harder if you have 21st century English and German in 0AD (attested somewhere). Showing they are relatives, almost impossible to imagine they could not with 2500 years divergence (or 2500 of divergence in one language and 3000 in another)…
Historical linguists don’t need to reconstruct ancestor to demonstrate genetic relatedness. Never been a required standard. Showing that lexicon has very high numbers of numerous correspondences in basic core vocabulary does not actually require you to be able to reconstruct how they are derived from the earlier forms through systematic sound changes – that *could* strengthen the argument that two languages are related, but sheer volume tends to be sufficient (or its absence sufficient for proving a lack of recent relatedness, without *extreme* sound changes).
Re; numeral systems, often have a strongly areal character in their form, e.g. WALS – https://wals.info/chapter/53, https://wals.info/chapter/54, https://wals.info/chapter/55, https://wals.info/chapter/89. Now systems can be the same without sharing lexicon and lexical sharing I can’t find an easy reference for. Quantifying lexical replacement in % of languages is very difficult (for’ex does the MSEA area constitute >1% of all languages on earth? Possibly? Certainly by speakers, though maybe not by volume given the surfeits of languages in the Americas and New Guinea).
@ Alberto
what do you make of Lusitanian ?
Wodko’s recent book seems revealing – “explaining the presence of IE traits in Lusitanian by prolonged contact with Celtiberians”, as explained by Mikhailova. I might obtain it
So I would agree; it seems that IE arrived very late in Western Europe
Also; wrt inscriptions; we may note that Thracian or Illyrian cannot he understood by way of Albanian
@Matt
The reconstruction of a putative Proto-Basque-Iberian seems to be the only way of proving a genetic relationship, or so I understand. Lexicon (which is mostly what we have, not much morphology) is more easily to take as loans, though of course the sheer volume you refer to would be enough (but then sheer volume of words is something we lack, not only, and obviously, in Iberian, but even in Basque.
But probably some background is needed here. When Eduardo Orduña (2005) first proposed the no well known equivalences between the Basque and Iberian numerals, he clearly stated that this was the result of Basque borrowing the numeral system from Iberian (since he didn’t believe in any sort of relationship between both languages). Ferrer i Jané (2009) elaborated on the study of these equivalences, and kept a more neutral position regarding the nature of the relationship between the languages.
Meanwhile, Javier de Hoz (2009), opposed this hypothesis as being implausible given his own research that had proposed that Iberian was only a vernacular language in SE Iberia, while in other areas (like Catalonia) it was used in inscriptions and as a lingua franca. Thus, the geographical distance and lack of any sort of direct contact between the Iberian speaking area and the Basque/Aquitanian speaking one made a borrowing impossible.
Joseba Lakarra (a vascologist, not an iberianist like the former ones) wrote an extensive critique too (2010) where he also endorsed the impossibility of a borrowing referring to de Hoz but adding his linguistic point of view from historical Basque, which made the equivalences unsuitable for being a loan. For example, the old presence of initial h- in Basque (i.e, not a modern development) which would not explain the addition of it in the case of borrowing the numbers 3, 10 or 20 (which lack the initial aspirate in Iberian, but have it in Basque), or the modern development in Basque of the loss of a consonant -n- in the case of the number 6 (sei in modern Basque, but apparently a recent development from the old sehi and the older seni, while the Iberian form is śei).
Lakarra’s main point here was to prove that the loan was implausible for both geographical (which probably include the cultural/archaeological too) reasons (not known contacts/interactions) and linguistic (not consistent with historical Basque).
As he puts it:
“4. Llegados a este punto, se me ocurre que deberíamos optar entre el parentesco lingüístico «duro» —que, como decimos, nadie parece animado a probar con los métodos estándares de la lingüística comparada— y la inexistencia de similitudes reales y significativas entre ambas lenguas; dicho de otro modo, las similitudes entre los elementos del (supuesto) sistema de numerales ibérico y los elementos (reales, documentados y conocidos por todos —empezando por varios millones de hablantes nativos los últimos 500 años—) del vasco serían no voces que, por lo que sea, los lingüistas no han conseguido convertir en cognados y en prueba definitiva del parentesco, sino puros espejismos debidos (no sé en qué porcentaje) al uso de determinado sistema de reconstrucción de los numerales ibéricos y a la voluntad manifiesta de querer creer en la existencia de esas similitudes pero sin afrontar la molestia de abordarlas desde un tratamiento comparativo estándar en lingüística histórica.”
My translation:
“4. At this point, it seems to me that we should choose between a “hard” genetic relationship -which, as we say, no one seems to be interested in proving through the standard methods of comparative linguistics- and the non existence of real and significant similarities between both languages; in other words, the similarities between the (presumed) elements of the Iberian numeral system and the elements (real, documented and known by everyone -starting by several million of native speakers in the last 500 years-) of Basque would not be words that for whatever reason linguists have not been able to turn into cognates, but a pure mirage due to (I don’t know in what percentage) the use of a specific system of reconstructing the Iberian numerals and the obvious will to believe in the existence of those similarities without making the effort to work with them in a standard comparative way in historical linguistics. ”
(The paper, for anyone interested, can be found here.)
However, the situation has changed since, when Orduña (2011) stated that he had changed his position regarding the nature of the relationship between Iberian and Basque, opting for a genetic relationship. A position that has only been strengthened in the subsequent years, and endorsed by, for example, Francisco Villar, and Indo-Europeanist who came from the side of rejecting such a relationship.
To my knowledge (but I don’t follow this so closely, so I may have missed it – if anyone knows more about it his contribution would be welcome), no further critiques from Lakarra or de Hoz have been published, and I don’t know their exact current position.
So, as you see, you find yourself in a rather curious position of defending the lack of a genetic relationship between Basque and Iberian and accepting the correspondences in the numerals but arguing that they are loan. A position that no one else (from either side) supports, since all reject the possibility of a loan as implausible for a number of reasons.
And to be honest, I’m still a bit perplexed as to why you seem to have this strong position on a subject that seems rather alien to you (though I could be mistaken), when my own position is that:
– We don’t know with certainty if Basque and Iberian are related or not. Not me, not anyone.
– The latest research (last 10-15 years) has made a significant progress in learning about it and the specialists involved (plus objective observers) are leaning towards a close genetic relationship between both languages, though they admit it’s still a long way to be proved.
– My 2 cents (or more like 1 cent) has been to highlight the latest research in ancient DNA, since this is something unknown by the authors working n this field. Now that we now fairly well the genetic/population history of Western Europe, the idea that these historical populations (Aquitanians/Basques and Iberians) could speak a closely related language is much more easy to explain than ever before (when Basques were thought to be some unique relict from the Paleolithic or any other weird -from out current perspective- theory about them) and Iberians were speculated to be a “Mediterranean” population unrelated to them.
I wrote a post where I mentioned this (it was not the main subject of the post) because I know that this research being recent and only (or mostly) available in Spanish, so it was meant to be informative about the current state of affairs. And since then I’ve been answering about it to further clarify things that seemed to me to be not well understood. Now, I’m not a specialist in the subject, and at this point I should say that if you are, then it would be more productive that you discussed it with the relevant people, not me. And if you aren’t, you may want to take what is informative rather than argue against it with me, because it makes no sense and I’m not even the right person to keep pointing out misconception or just lack of information for debating about it.
@Rob
I haven’t read Wodko’s latest book about Lusitanian. It’s another difficult case as with all the poorly attested languages. I don’t have any strong opinion whether it’s a Celtic language with some late influence from Latin, a pre-Celtic ones (para-Celtic, given its similarities in any case), or if it’s just another non-IE language with some strong Celtic (and weak Latin) influence difficult to uncover due to being late attested.
But overall the late arrival of IE to Western Europe seems quite uncontroversial with the current data.
http://euskararenjatorria.net/wp-content/uploads/2015/07/EL-IB%C3%89RICO-LENGUA-USKEIKA.-SUBSTRATO-DEL-ESPA%C3%91OL-Y-PATRIMONIO-DEL-EUSKERA-III.pdf
Hola Alberto, esta es una tesis doctoral muy buena que puede ayudar a entender la estrecha relación entre Íbero y vasco- Tanto los vascos como los ´´Iberos y Tartesios de la edad del Hierro son descendientes directos de los Campaniformes Ibéricos- Si tienes tiempo para leerla seguro que te resultará interesante – El vasco-Iberismo es actualmente la teoría más aceptada aunque es obvio que existen opiniones para todos los gustos. Un saludo
@Alberto, my impression above was that you seemed to be un-familar with / confused / glossing over the distinction between 1) “reconstruction” and 2) providing sufficient evidence for a genetic relation between two languages.
The former requires detailed reconstruction of stage of change in morphology and phonology, the latter only requires a large number of regular correspondences across a large number of categories of basic vocabulary (that which is unlikely to be borrowed) and morphology and typology. A large volume of basic lexicon is impossible to be borrowed (whatever lexicon ultra-sceptics seem to believe – this is empirically demonstrated to be resistent to borrowing even in extreme cases, or certainly no less resistant to borrowing than shared morphology). Reconstruction of proto-Indo-Iranian or proto-Indo-European was never necessary for identifying that Sanskrit and ancient Greek were both IE languages!
(If you were familiar with the distinction, I thought it would be useful to give you the opportunity to demonstrate it, as otherwise anyone with a basic linguistic knowledge would probably be very unimpressed!)
If you want to make claim that the corpus of Iberian inscriptions lacks enough basic vocabulary to make this comparison, that is fine and could be bearable (I do not have an expert knowledge of the Iberian corpus, though I would guess neither do you). But to argue that it we do have such a corpus of basic vocabulary but shared basic lexicon is only visible through numerals seems impossible, whatever the argument about implausibility of borrowing numerals on linguistic grounds. (While those geographical arguments about lack of contact don’t seem to make much sense to me since there is clearly borrowing of words relating to trade, urbanism and commerce. Pretty impossible if they never met… To argue lack of contact rules out loans, we’d be in a strange situation of having to deny the existence of *any* loans between Iberian and proto-Basque, which position is obviously not sustainable.)
Happy to leave this conversation here! No need for you to reply.
@Matt
“If you want to make claim that the corpus of Iberian inscriptions lacks enough basic vocabulary to make this comparison, that is fine and could be bearable (I do not have an expert knowledge of the Iberian corpus, though I would guess neither do you).”
And I was pretty sure that we both knew well enough the limitations of the Iberian corpus, not only because I clearly mentioned it in the original post and because I have been repeating it ad nauseam (and really, I don’t know how this hasn’t got through to you), but because I thought it’s a more or less obvious thing that is not particular from Iberian, but from basically all the ancient languages only attested in a few inscriptions made in stone.
And I did also in your first comment about this back then told you (without any intention of being rude) that you were overestimating your ability to judge this matter, and since then I’ve just been clarifying (or trying to) your poor understanding of the situation. Just to listen now that you wanted to give me the chance to show that I’m not confused about some very basic linguistic concept?
Come on, Matt. I’ve been polite and patient with this, first for the respect I have for you and second because I thought it was in everyone’s interest that you didn’t keep spreading misinformation about it. Given your last comment where you still insist in knowing more than all the experts, I do have to give up hoping that we’ll still have more productive exchanges in other subjects.
@Gaska
Gracias, no había visto tu mensaje porque se había quedado bloqueado por el filtro de spam (quizá no le gustó el link). La leeré en cuanto tenga oportunidad. Un saludo!
Alberto, I have been reading this conversation and I wanted to say that the discussion regarding Ibero-Basque is “genetically” overcome. The Basques are a kind of isolated Iberians in the mountainous areas of the Pyrenees- There is no greater mystery about it. On the other hand, the Basques and the Iron Age Iberians are identical to the Iberian BBs not only in their autosomal composition but in their uniparental markers-The genetic continuity between 2,500 BC, the Iron Age and the present, is so evident that no one can deny that there is also a linguistic continuity, and what we have at the end of those periods?- Basque, Iberian and Tartessian, three non-Indo-European languages. Then the conclusions for me are obvious
1-Neither BB culture nor P312 spoke IE languages, because there is no reason that could make us suspect that there were changes in the language. Or do you know any explanation that might be reasonable?
2-All the Iberian cultures (Bronze Age) are overwhelmingly R1b-P312 / Df27- Las Cogotas, Las Motillas, Bronce Valenciano, Bronce del Guadalquivir, Bronce Atlántico, El Argar, and all of them are archeologically and culturally linked with the historical peoples of the Iron Age (except obviously the Celtiberians who were the ones who introduced the Celtic into the peninsula)
3-The link between Basque and Iberian is more than evident, not only in the issue of numerals, but that there are hundreds of Iberian words that are identical to Basque (we do not know if they mean the same, but can help translate many texts )
4-We have the doubt to check the genetic composition of the western Iberians, that is, Galaicos, Astures, Vacceos, Carpetanos, Lusitanos y Vettones. There is a very interesting project for vacceos and vettones because we have managed to recover a lot of skeletons of children in the villages (They practiced cremation in certain cases, also they left the corpses of the warriors without burying them to be devoured by the vultures, wolves… but the children who did not reach two years were buried under the floor of the houses- With total certainty (we have already advanced some results) they will also be Df27, with which the linguists will have more work trying to decipher the languages spoken by these peoples
In short, we have what the Kurganists dream of having, that is, a clear genetic link
between a language (Iberian/Basque/Aquitanian) and a specific genetic marker (R1b-P312/Df27)- that is, for us the game is over, because this is the only way to scientifically demonstrate the origin of a language
Can you imagine that R1b-L51 / P312 would have appeared in the Yamnaya culture? – Everyone would have said that the steppe theory was correct, that is to say that IE extended to the West and East thanks to the massive migrations of R1b and R1a from that culture. However, things have not gone as the Kurganists expected, and it turns out that in Yamnaya there is only R1b-Z2013, a lineage that in no case can be related to historical IE languages. The ridicule they have done is sidereal, because the Mycenaeans and Hittites are currently J2 and the typical lineages of the steppes have never been found in Western Europe
The Kurganits only have the “ghost steppe ancestry”, who does not know exactly where it came from or what samples they have chosen. Before it was Yamnaya ancestry, now steppe related ancestry, before it was EHG-CHG, now it also has EEF and WHG etc, Yamnaya culture has become the Yamnaya horizon, chalcolithic has become eneolithic steppes……..
I belong to a Spanish Foundation for the protection of Hispanic culture (Iberia and Latin-América) and we are concerned about how all these “genetic events” are developing, Thousands of unpublished samples, use of those samples that interest us to demonstrate our theories, manipulation of public opinion, risky and unscientific conclusions in many of the published papers, that is to say the opposite of how a serious and rigorous scientific debate should develop- We never imagined that collaborating with foreign scientists was going to become a kind of nightmare- The lesson is learned, many geneticist, archaeologists and students are aware of the danger of leaving all these ancient samples to the hands of people who do not know the Prehistory of Spain without any control,
so, papers and doctoral theses are being prepared using truly interesting sites (the first will be Los Millares, La Pijotilla and Marroquies Bajos) that span crucial years in the Spanish Chalcolithic-
Te cuento esto como español porque supongo que podrías estar interesado en colaborar con nosotros de manera independiente, especialmente en todo lo referente a la utilización de herramientas informáticas para modelar ancestrías, componentes autosómicos etc. Como creo que ahora he utilizado el enlace correcto puedes ponerte en contacto con nosotros en mi correo electrónico.
@Gaska
It would be difficult to disagree with your analysis of Western Europe, but what do you make of the R1b-L51 diffusion into the Italian peninsula? IIRC Vennemann grouped Ligurian with the indigenous Iberian languages, but I’m not really familiar with his arguments.
@Marko
The Etruscan plots near the Iberians, and descend directly from Northern Italy BBs (Parma, Olalde, 2.018) and the Villanovan culture- We only have three samples, one is an African outlier and other has in my opinion an Illyrian marker that has also been found in the Nuragic culture and in Croatia. Then an Etruscan-Balkan connection seems obvious. Regarding the Italics they are very similar to the Etruscans and R1b-U152. I think they entered mainland Italy at the beginning of the Iron Age – I hope that more information about the Etruscans will be published soon. If they turn out to be P312 then it will be the nail in the coffin of the steppe theory as it has been interpreted since Haak 2015-
@Gaska
If the rumors are true, the samples should mostly belong to R1b/I1. The problem to me then is the fact that Etruscan is unlikely to be genetically related to the Iberian languages, which would weaken the R1b-L51/Beaker = ‘Vasconic’ hypothesis somewhat.
@Gaska
Yes, I think that things are coming together in the Basque-Iberian front, and with the current linguistic and genetic data is difficult to actually argue against them being genetically related. As it’s become difficult to argue that Bell Beakers brought Indo-European languages to Western Europe and Italy, given that all the evidence we have is against it (and yes, as you also mention, the amount of evidence against the steppe people being Indo-European as a whole has only been growing and a theory about the origin and spread of IE languages cannot rely on a set of very unlikely events at every stage. But let’s be patient and wait for more definitive data about this).
@Marko
I think the situation in Italy is quite more complicated than in Iberia. It’s going to require some very good sampling (as the Balkans) to be able to draw conclusions.
Regarding Etruscan I really don’t know anything special about the language itself. I noticed that FrankN referred to some quite specific features that apparently are present in Etruscan, Hurro-Urartian and NE Caucasian languages (namely case stacking and antipassive voice, both of which are present in Basque too). This might be a coincidence; I really have no opinion about it. But I’ve been intrigued by the possible Basque – NE Caucasian relationship for a while. If the latter comes from Hurro-Urartian, it’s going to be interesting to get samples from this population (which I’ve speculated to be the best candidate for the late Yamnaya/Catacomb intrusion in West Asia crossing the Caucasus). Dagestan is also a place where the original steppe language could have survived better than anywhere else (and NE Caucasians are among the most direct descendants of Yamnaya people, if such thing exists). All of this is just thinking aloud, not any sort of hypothesis.
Overall, about the language of Bell Beakers or at least about pre-IE Western Europe I’m looking forward to what linguists can make out of the new data we have. As I explained in another post, the mainstream among Spanish linguists in the last two decades (maybe equivalent theories in other countries) has been that IE was very old in Iberia, while non-IE would have been later arrivals (either with agriculture or at some later point). However, this is incompatible with what we know today, and places that were non-IE until the roman conquest can’t have any IE substrate. I used a paper about Catalonia as an example, where only 10% of the name places could be found to be Iberian, while 50% was an unknown IE (0% of it Celtic related), with Greek and Latin counted separately too.
We’re obviously talking about pseudo-IE substrate. The main reasons to explain it are that: 1) No correspondence has been found for them in the Iberian inscriptions that we have (which is nothing strange, given the limited corpus available) and 2) that similar roots have been found around Western Europe (mostly) where IE languages are spoken (and not Iberian), with some often dubious IE etymology.
The reality is that said 50% name places must be actually Iberian, even if we don’t have proof of it in the available inscriptions. Once this is recognised and accepted it will start to yield some interesting results when compared to the rest of Western Europe, finding many coincidences (no need to actually find them – they’re already known, though qualified as IE).
With carefull analysis, I’m hoping that in the next decade we’ll be able to figure out the linguistic landscape of pre-IE Europe and maybe be able to tell what came from Anatolia and what from the steppe. A difficult task, but also a fascinating one for linguists and for the rest of us to follow.
Alberto
Not much of the stuff on Vasconic in English; it is either old (eg Ice Age relict theories) ; or inconsistent with the facts (eg Koch).
Rob, yes, it’s unfortunate, but given that the Ibero-vasconic academic research has only taken off in the last decade (before it was mostly in the amateur realm) and that it’s still a work in progress it will take a little while before we start to see anything published in English.
I hope that the renewed interest that ancient DNA has brought to the Bell Beaker Culture (as the base of Western European populations, while before its importance was quite unknown, but even the most optimistic theories wouldn’t have predicted what we’ve seen) will also bring a strong interest in researching their original language. And this will inevitably have to involve Basque, apart other ancient Western European languages now extinct. It won’t happen overnight, but throughout the next decade we will probably see much more on this.
@all
Have you read Bengtson for some newer work on this particular subject? He’s a proponent of the genetic relationship between North Caucasian and Basque, and posits a vast substratal area for Western Europe, with the substrate influence being most pronounced in Celtic as had already been laid out by Venneman (Bengtson cites Kassian who contends that the latter has a significant number of Swadesh words of Vasconic origin, and speculates that Celts are ‘Basque-like people’ who underwent massive language shift to IE). He posits a seperate origin for Etruscan if I’m not mistaken.
In that vein it would be interesting to see whether anything can be learned from Hallstatt archaeogenetic data. The early Hallstatt Iron Age kurgans are quite distinctive, so it would be useful to test whether the buried are in any way genetically differentiated from the preceding population. Thus far we only have the two samples from the Czech necropolis, and an Y-DNA from an Austrian Hallstatt chief.
@Marko
Yes, the Vasco-Caucasian hypothesis has been recurrent and has always been intriguing. It’s a pretty difficult question because even in the best case scenario (both languages being related at a lower time depth then the Early Neolithic by the way of the steppe migrations in the LN/BA) it’s still a 5000 years split of two very isolated languages. However, *if* we get to the point where it’s clear and widely accepted that the steppe was non-IE, it will make sense to start looking for the possible languages that they spoke, and I guess there are basically 3 prime candidates (Basque-Iberian, Uralic and North-East(and West?)-Caucasian.
Bengtson has indeed a recent and very comprehensive (500+ pages) work on the matter that is freely available here:
Basque and its closest relatives: A new paradigm
“In direct contradiction of these kinds of statements [the uniqueness of Basque], the thesis of this book is that Basque is demonstrably related to other languages, i.e., that a scientific analysis of the evidence leads to the most probable conclusion that Basque is, at first remove, most closely related to the North Caucasian language family.”
Still, we must go step by step. First we need to solve the IE question. Then everything else (Uralic, Basque, Caucasian families, Etruscan, etc…) will have a much more clear ground to start working on.
EDIT: I should have mentioned the other work you refer to about the substrate in Celtic even though it’s in French:
Confirmation de l’ancienne extension des Basques par l’étude des dialectes de l’Europe de l’Ouest romane
@ Alberto
I lean toward the possibility of the steppe spreading at least a couple of languages. PIE from a steppe – MNE zone, and other from steppe-Caucasus zone. The latter could explain Vasco-Caucasian. Curiously, in one of his talks, Bengtson states that Rene Lafon suggests that Vasconic is not native to western Europe, but arrived during the Copper age in the latter half of the 3rd millenium.
@ Marko
”In that vein it would be interesting to see whether anything can be learned from Hallstatt archaeogenetic data.”
Imaginably it would confirm the long-duree process of ~ Urnfielf Halstatt La Tene, with extra nuance, of course
For ex, as classically envisage – La Tene periphery taking over formerly Western Halstatt chiefdoms, with associated genomics shifts from C to NW Europe, fixation of R1b-P312, and ? langauge shifts
@Rob
Yes, the process might have been rather complex, and I’m not sure what should have enabled the Celts to effect large-scale language shifts without replacing the native population. If true, they must have already had a rather complex political organization. That’s why Y-DNA from the core Hallstatt territory would probably be informative. There’s one G2a-L497 from the kurgan of a chieftain at Mitterkirchen, Upper Austria, which should be close to the epicenter of early Hallstatt regions. The more peripheral man from the Bylany necropolis had R1b-U152.
@Rob
Yes, that’s a possibility. the steppe R1b-L23+ groups don’t seem to have been IE either when going to Western Europe or when crossing the Caucasus and appearing in West Asia. However, the steppe cannot rely on R1a-M417+ groups alone for the spread on IE languages. It wouldn’t explain Greek and probably other language from the Balkans, Italic, probably Armenian,… So all these ones would require a MNE populations to have been PIE too (which I think it’s what you meant).
To clarify this a bit we’d need to solve to pending questions: the specific origin of the Bell Beaker (R1b-L51) people (whether it’s related or no to the CWC people) and the specific origin of the Sintashta people (whether they derive from the forest steppe preceding groups -and ultimately share the origin with CWC- of if they are recent migrants from East-Central Europe that could have acquired a new culture and language different from the earlier steppe groups).
The rise in frequencies of the European variant of the Lactase Persistence allele had been associated with population movements and possibly language spread. However, aDNA has been disproving this hypothesis in the last few years. Here’s an image that summarizes it quite well:
Ian Mathieson twitter
The question is: where did they get the details about the Indus Valley? Unpublished samples or just from the few published ones?
“The question is: where did they get the details about the Indus Valley? Unpublished samples or just from the few published ones?” — Alberto, i think it’s from the few published ones, Ian writes here
http://mathii.github.io/2019/10/12/the-spread-of-the-european-lactase-persistence-allele
He writes — ” Looking at the data from Narasimhan et al. 2019 it seems that the allele appeared in South Asia much later than in Europe. Sampling is a bit limited, but its earliest appearance in data is around 2000 BP in Butkara and Swat. The present-day frequency of ~25% in some South Asian populations (e.g. 1000 Genomes PJL) suggests strong, recent selection perhaps similar to what we see in Iberia.”
I wonder if theory of ahimsa and non-violence(non-vegetarianism) that was widely propagated post 300 BCE especially in north india contributed to this selection.
Btw, since this is a topic about Iranian ancestry: have any of you taken a look at the heavily ‘Iranian’ Bronze Age genomes from Ashkelon? One of them has Y-DNA L, too, and they’re dated to the period which corresponds to the Mitanni/Maryannu takeover of the Levant.