There was a new paper published earlier this month that is really interesting for understanding the human dispersals in the Americas. Of course, with the help of some newly sequenced aDNA. From the abstract emphasis mine):
“We sequenced 15 ancient human genomes spanning Alaska to Patagonia; six are ≥10,000 years old (up to ~18× coverage). All are most closely related to Native Americans (NA), including an Ancient Beringian individual, and two morphologically distinct “Paleoamericans.” We find evidence of rapid dispersal and early diversification, including previously unknown groups, as people moved south. This resulted in multiple independent, geographically uneven migrations, including one that provides clues of a Late Pleistocene Australasian genetic signal, and a later Mesoamerican-related expansion.”
I can’t comment in details about this very good paper that should satisfy those who are more into the history of the Americas than I am. So to those I can only recommend them to read the paper and enjoy it.
Here I’ll just comment briefly on a few details that are mentioned in the abstract excerpt above and that are somehow linked to some comments from a previous post about the population history of Northern Siberia (Sikora et al. 2018).
But first, regarding the mention of those two “Paleoamericans”, I’ll just say that they were samples that had been speculated to be from a different population due to their different cranial morphology compared to other Native Americans. However, the genetic analysis shows that they’re genetically the same as the rest, showing that cranial morphology can vary due to drift without any admixture involved.
The two other things I want to comment about are those related to the “previously unknown groups” and their relationship to that Australasian signal and to a Mesoamerican distinct one.
So about the Australasian signal, this is something that had already been found before (Skoglund et al. 2016) and that was specially showing up when comparing the affinity of Suruí (and Karitiana, though a bit less) to Mixe (and Pima, but a bit less) to Australasians (from Andaman Islands and Papua New Guinea).
My observation from the previous post about the Siberian paper was that the relationship between Anzick-1 (a ~12.5 kya sample) and Suruí/Karitiana to Ancient North Siberians (ANS) was symmetrical and it didn’t seem that there would be anything significantly different about those populations from the Amazon. Instead, I found that it was Mixe and Pima the ones who had a different relationship to ANS compared to other NA, showing some admixture from an unknown source (non-Eurasian in any case, since they didn’t show any increased affinity to any Eurasian population.
However, here they compare with D-stats directly Anzick1 and Suruí to Australasians, and from figure 4A it looks like D(Anzick1, Suruí; Australasian, Yoruba) has a Z-score of about -2. It does not reach significance, but it’s there. Interestingly, the signal is also found in a ancient samples from Lagoa Santa (SE Brazil) dated to around 10.4 kya. Using qpGraph, this Australasian admixture is estimated to be around 3% in these samples (and modern Suruí). How it got there (if it’s real, that it well could be) is unknown at this point, but it’s important to keep in mind that it was already present at least 10.4 kya, so it cannot be from any kind of Holocene migration of Australasians to South America. The possibility is that there was a small (?) population of Australasian origin at the arrival of the migrants of NE Siberian origin.
The second thing is even more interesting: when on a similar D-stat as the above one, but with Mixe in the place of Anzick1, the result reaches significance (Z-score around 3 and a bit more in Fig. 4A). This is what had already been found before and used to argue about the signal being real and strong. However, while I don’t doubt that the signal is there, what they had failed to realize was that there is actually something specific in Mixe that makes this stat significant. And what it that? Here’s what the paper says about it (emphasis mine):
“by fitting f-statistics-based admixture graphs, and found that even though Anzick1, Spirit Cave and Lagoa Santa are separated by ~2000 years and thousands of kilometers, these three individuals can be modeled as a clade to the exclusion of the Mesoamerican Mixe (13). While we did not find evidence rejecting this clade using TreeMix and D-statistics (13), further SFS-based modeling indicates that Mixe most likely carry gene flow from an unsampled outgroup […] Hereafter we refer to that outgroup as ‘Unsampled population A′ (UPopA), which is neither AB, NNA or SNA [Ancient Beringians, Northern Native Americans or Southern Native Americans], and which we infer split off from Native Americans ~24.7 ka, ranging from 30-22 ka (95% CI; this large range is a result of the analytical challenge of estimating divergence and admixture times in the absence of UPopA genome data).[…] Under a model with a pulse-like gene flow, we inferred a probability of ~11% gene flow from UPopA into Mixe ~8.7 ka (95% CI: 0.4-13.9 ka; the wide interval potentially reflects unmodeled continuous migration)”
I don’t have the tools and samples to test these things formally, but from the data provided by the paper I’d propose a slightly different interpretation. For what I see in Figure 4A, it’s clear that Mixe seems to have admixture from an outgroup too all the populations tested (see for example these 2 below):
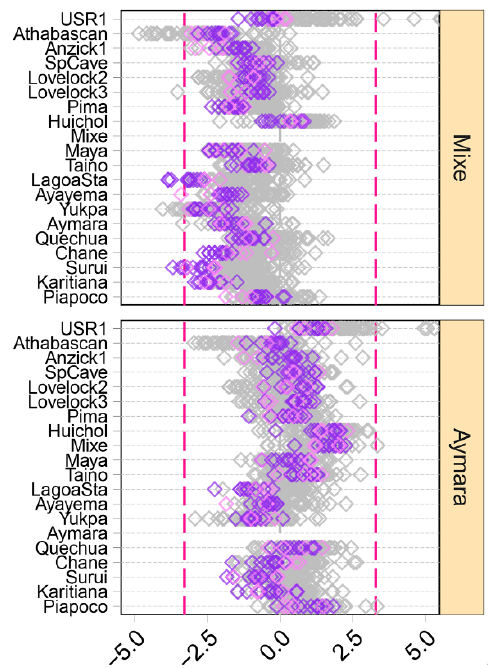
The shift that we see to the left in the case of Mixe basically shows that while the pattern of affinities is the same as in Aymara, the number of shared alleles with all the Eurasian populations represented by the diamonds is lower than Aymara’s shared alleles. The question here is what kind of population would be an outgroup to Native Americans. And the first answer is simple: roughly anyone else. True. But then we could find the specific population in Eurasia (modern or ancient) with which Mixe would share more alleles than other NA groups (especially those without Mixe-related admixture, like Suruí). However, we don’t have such population. When they say “unsampled”, they mean unsampled. So no, we’re not talking about potentially “anyone”, but quite specifically about a population that does not only seem to be an outgroup to NA, but also an outgroup to Eurasians.
If this is true, then who can be an outgroup to Eurasians? Basically there are 4 options:
– An African population (meaning a population that went Out of Africa after the main OoA event that gave birth to most Eurasians, and that somehow reached Central America some 9 kya). This one is the least likely, really.
– An early OoA population (meaning a population that went OoA before the main OoA event). We know from archaeology (and with some support from genetics) that such early events did occur but they hardly contributed to later Eurasian populations (maybe a tiny bit to some SE Asian/Australasian populations?). So this one would mean that such population made it to the Americas and survived somewhere around Central America until the second major wave arrived.
– Archaic hominins. Like Neandertals or Denisovans. A small admixture from such groups (on top of what other NA already have) would make the shared drift of Mixe with all other AMH be lower. So is it possible that some form of archaic hominin lived in America and survived until some 9 kya?
– A fourth option would be an early “generic” Eurasian population, something similar to Ust-Ishim samples from 45 kya, but not directly related to Ust-Ishim. This, to me, seems the most realistic of the options. We know that such populations (part of the main OoA) reached Siberia quite early (as Ust-Ishim shows), so why couldn’t some of them reach America and survive till the second bigger wave from around 17 kya? Archaeology has found small evidence of human habitation in America from around 50 kya (IIRC), so that could be it. For me the question in this case (but also in whichever of the previously mentioned cases) is why didn’t such population thrive in America in a similar way as the later one did as soon as they arrived?
Whatever the truth turns out to be, a very interesting question the one this paper has opened with the UPopA admixture into Mixe and other Mesoamerican populations (Huichol, it seems), though not limited to them, since there has been an expansion of these Mixe-related populations that has made that the admixture from UPopA is present in many (most?) modern NAs, just to a lower degree (in any case, keep in mind that as in the case of the Australasian-related admixture in SNA, we’re talking about a very small genetic contribution to the autosomes and none to either the paternal or maternal lineages, so the contribution itself is mostly anecdotal. What is important is to know if any of those populations were in America before actual NAs arrived).
UPDATE:
As pointed out by Frank (personal communication), and in line with his observations about the Kennewick Man in the previous post about the population history of NE Siberia (see comment here) , on Figure 2D we can see that Kennewick Man already has Ancient Beringian admixture shown by the extra East Asian:
I confirmed this by using Global 25 datasheet and modelling Kennewick as a mixture of Clovis (Anzick1) + something else:
Kennewick
Clovis 82.8%
Chukchi 8.5%
Ulchi 7.7%
Papuan 0.4%
Eskimo 0.2%
Australian 0.2%
Yoruba 0.2%
Dai 0%
Atayal 0%
Han 0%
Onge 0%
Ust_Ishim 0%
Iberomaurusian 0%
Ju_hoan_North 0%
Distance 2.1449%
We can clearly see the extra NE Siberian/Beringian admixture there.
Unfortunately, trying a similar approach to detect any admixture in either Suruí or Mixe didn’t work, both coming out as 100% Clovis. A more sensitive test should be performed with these ones (well, it’s already done with Suruí, both in this paper and in Skoglund et al. 2016 showing the Australasian related admixture). For Mixe, maybe someone would like to try some of the options outlined above, like:
D(Chimp, African; Anzick1, Mixe)
Where African could be from Iberomaurusian to Ju Hoan.
D(Chimp, Archaic; Anzick1, Mixe)
Where Archaic could be Neanderthal or Denisova.
D(Chimp, Ust-Ishim; Anzick1, Mixe)
And see if any of those give a Z score of around 2 or greater.
Early human dispersals within the Americas – Moreno-Maya et al. 2018


Do you see any possibility of West Eurasian pre-Viking contact from any of the data so far? I would imagine such material would be censored due to the rather extreme political sensitivities , but I’m curious anyway. What about Genetiker’s analysis of the Chinchorro remains?
I ask about this potential influence, because I KNOW that it existed, based on evidence in this link – and the evidence really is basically overwhelming. Heyerdahl wasn’t some madman, you know…
https://justpaste.it/4tqjz
Yes, it might sound wacky, maybe even racist (and I honestly want nothing but the best for humanity as a whole, regardless of racial differences, as we are all basically the same), but it just happens to be the likely truth. Of course though, me saying it is definitely true does not necessarily make it so, so I’m wondering what your opinion about this is. Please look at that link though, it’s a treasure trove of information.
Many thanks if you remain open-minded, and don’t reject evidence just because it seems so fantastical (which I don’t see why it should, the idea of Oceanic influences is much more impressive considering the size of the Pacific and nobody denies that fanatically).
Jacob
@Jacob
I don’t see why the presence of West Eurasians in America before the Vikings would be too controversial, let alone racist. Or why such information would be kept hidden.
My only problem with it is that I don’t see much evidence so far. Nothing in the aDNA record from America (and we’re getting a growing collection already) nor in the modern one shows any such admixture. If those West Eurasians arrived to America (something difficult but possible, like many other feats of the past), where did their DNA go? The Chinchorro mummies had contaminated DNA if I remember correctly. We’ll need better samples to convince anyone about such presence. If the lack of any traces of it is not enough evidence of absence already.
I do keep an open mind, and if such DNA appears it will be great. But so far, there’s nothing about it.
@Alberto Well the connections that can be made basically put these West Eurasian explorers down as being the elite rulers, and then of course claims can be made that things like smelting technology and higher civilisation were spread by these people (there are all sorts of (mainly South) American and Polynesian folk stories about this, everything is contained within Thor Heyerdahl’s works). This is the stuff of Stormfront’s dreams, and indeed that link I posted was from one of Genetiker’s blog posts about “The White Gods”. There’s obviously a lot of room for censorship here – not least because the guy doing a lot of this work is Eske Willerslev, who seems like a typical crazed “protector of the weak” Scandinavian (and this is very clear from some of his interviews with the press).
The Spirit Cave mummy that was recently sequenced blatantly had Caucasoid features, there’s no ifs or buts about it. Ignore the reconstruction and just look at the skull: http://www.friendsofpast.org/img/06SC/06SC5lat.jpg – if you know anything about skull shapes, it should be pretty clear (and yes, skull shapes only have so much accuracy, but when comparing completely different races like Native Americans and West Eurasians its accuracy is extremely high). Add on to that that, just like those images in the link I provided, his remains also preserved some of his auburn hair and even some of his skin, which was appropriately light. I genuinely believe that, if the results do in fact show that he had no West Eurasian admixture, they were forged. All the evidence from what we know about his remains points toward him being similar to the North American Si-Te-Cah people, who, according to the Paiutes, were “red-haired giants”, and the Paiutes even passed locks of this red hair down from father to son. These many stories across the Americas of parallel plot don’t count for nothing – they all point to the same thing.
The fact is, that we don’t even need aDNA to confirm this idea of West Eurasian pre-Viking contact, because of everything in that link I sent. The remains are so clearly Caucasoid, it is cognitive dissonance to claim otherwise. Yes, aDNA is more precise, but they can also be tampered with – not least given the absolutely ridiculous concentration of power that exists in the aDNA community (basically two labs control almost all of what is published).
And to stoke the flames here: what about the Swastika? Did these Native Americans just happen to adopt that symbol by chance? There are basically infinite simple geometric forms to choose from, why choose that? Just a coincidence, right? And before anyone dares to play the White supremacist card – I’m legitimately of fully Ashkenazi Jewish descent, which doesn’t really matter, but also does matter in this twisted modern-day world of accusations and labelling.
I can only hope that the truth isn’t destroyed.
Jacob
Apparently, the Paracas skull (same red hair as all those examples I posted, but extremely elongated) had mtDNA U2e, which is without doubt West Eurasian and probably West Asian. Some progress at least.
There is also the whole mtDNA X in Native Americans thing, which is just so clearly West Eurasian too – I don’t care if the Native American subclade (X2a) is different to that of West Eurasians, you’d expect that given how long ago the divergence must have been.
Look at this map: https://www.ancient-origins.net/sites/default/files/styles/large/public/Genetic-Map-of-Haplogroup-X.jpg
Does it seem more likely that a clearly Middle Eastern branch that developed mainly in the Neolithic somehow made its way all the way across the Bering Strait without leaving so much as a trace in Siberia, or that it went West via the Atlantic?
Jacob
@Jacob
I’d prefer you didn’t post here about conspiracy theories and accusations of forgery of ancient DNA. This is not the way you will convince me or anyone else.
If a skull that you thought was West Eurasian has been sequenced and it has turned out to be Native American, then just accept that you were wrong (not you specifically, but the assumption itself). And by accepting this, you will see that the evidence you believed to be very solid suddenly becomes much weaker. This is the kind of power that aDNA has in separating the myths from the truth.
I’m sure that we would all find it really interesting to see West Eurasians in the Americas some 2-3 kya. Even the Native Americans would see their “legends” confirmed, and it would be proved they were not fantasies. So if that did happen, just wait, because sooner or later it will be found.
The case of mtDNA X2a is interesting. But again, the people who carry in significant frequencies this lineage don’t seem to have any “recent” West Eurasian admixture. Maybe when we find an X2a sample from 2-3 kya it will be different, who knows. But we have to wait for that to happen.
Have no doubt that the truth will be found and not destroyed. But be patient and stay open minded you too. It might turn out that your believes were wrong, and that’s ok.
A few other models using a sample from Chile as source instead of Clovis, who already shows up a bit of Siberian ancestry:
Clovis
Chile_LosRieles_10900BP 84.6%
Aleut 9.4%
USR1 5.6%
Koinanbe 0.4%
Distance 1.8046%
Mixe
Chile_LosRieles_10900BP 92.2%
USR1 7.8%
Aleut 0%
Koinanbe 0%
Distance 2.8544%
Pima
Chile_LosRieles_10900BP 87.2%
USR1 12.6%
Koinanbe 0.2%
Aleut 0%
Distance 2.8034%
Kennewick
Chile_LosRieles_10900BP 57.2%
USR1 39.4%
Aleut 3.4%
Koinanbe 0%
Distance 2.5649%
Surui
Chile_LosRieles_10900BP 100%
Aleut 0%
USR1 0%
Koinanbe 0%
Onge 0%
Papuan 0%
Distance 7.1597%
Karitiana
Chile_LosRieles_10900BP 100%
Aleut 0%
USR1 0%
Koinanbe 0%
Onge 0%
Papuan 0%
Distance 5.4686%
@ Jacob, Alberto:
I think the case of X2a is now almost settled. It is attested for Kennewick, which means arrival before 7 ky BC. The upper boundary is given by the X2a split time from the Eurasian X2, estimated at ca. 10.8 ky BC in Soares 2009. I strongly suppose X2a is connected to the “Beringian” element in Kennewick that now has become obvious, and that means trans-Pacific rather than trans-Atlantic early Holocene entrance.
Interestingly, while the study demonstrates strong genetic link between Kennewick and Na-Dene Speakers, linguistics suggests that the family is unlikely to have formed before ca. 4,000 BC. IIRC, a NaDene Arrival in the Americas around 4,000 BC was also the conclusion from the Sikora e.a. 2018 demographic modelling of Paleo-Indians, and the date estimated by Skoglund/Reich. There is strong indication of corresponding substantial population replacement: After ca. 4,000 BC, once widespread mtDNA D4h3a (Anzick1, Lagoa Santo) completely disappears from North America (both ancient as modern DNA).
Then, Sikora e.a. suggested another Beringian migration, not directly related to NaDene, and somewhat later than 4,000 BC, being responsible for Paleoeskimos. [For the temporal proximity, Skoglund/ Reich were still uncertain whether NaDene and PaleoEskimos reflected one and the same, or two separate immigrations].
Finally there is BigBar (5.3 kya. Inland British Columbia), for which Moreno-Mayr state “D-statistics of the form D(Aymara, NA; BigBar, Yoruba) and D(USR1, NA; BigBar, Yoruba) suggest Big Bar represents a previously undetected outgroup to non-AB Native Americans, one that diverged prior to the NNA/SNA split” (i.e. prior to ca. 13.7 ky BC).
In summary, the demographic history of the Pacific NW seems to have been much more complex than previously assumed, including numerous waves of ENA immigration, early internal splits (USR_1, BigBar), population replacements e.g. around 4,000 BC, relatively recent “Mesoamerican” arrival, etc.
As they say: “It was previously surmised that Clovis (Anzick1) and Western Stemmed (Spirit Cave) technologies (46, 56) represented “genetically divergent, founding groups” “Western Stemmed Technology (WST) makes early appearance in S. Chile, so for the Pacific Coast and the SNA Expansion, their findings match archeological expectations. OTOH, as already in previous studies, Anzick1 has once more been shown to relate to this population as well.
I have always been wondering if equating Anzick1 with the Clovis Culture is correct. Anick is an ensemble that in the West Eurasian BA would be labelled as “princely grave” – an infant buried with lots of finely-worked Clovis points. However, I doubt that Clovis was already so socially differentiated that it had “princes”. This more looks like a sacrifice – and the human infant sacrificed may well have been captured from a competing group, i.e. WST.
Intriguingly, BigBar corresponds to what would be expected from “ancestral Clovis” – early genetic differentiation, and presence along the supposed “ice-free corridor” instead of the Pacific coast. Moreover, in their PCA BigBar plots amidst non-Athabascan North Amerindians, essentially from EC N. America, i.e. where one would assume most of Clovis ancestry to be found (albeit the Holocene history east of the Rockies was also quite complex, including the rapid Algonquian expansion during the Woodland Period 3000 kya).
I’ll comment seperately on the Mixe stuff
About the unsampled population (UPopA) that has contributed to Mixe and other Mesoamericans:
First of all, let me thank you, Alberto, for the observation that “Mixe seems to have admixture from an outgroup to all the populations tested.” That is a great and very important observation.
I disaggree, however, on some of your conclusions and scenarios derived from it.
1. While they speak about “Eurasians”, their panel is effectively North Asians, East Baltics, plus Sardinians and French (see Fig. 4C). They systematically filter out French alleles in order to correct for a putative post-Columbus contamination (see Fig. 4B), thereby effectively depressing West Eurasian impact on the stats. However, Fig 4B shows that the extent of such filtering symmetrically affects the Mixe-Lagoa Santa relation, so we may conclude that any population that has contributed major ancestry to modern French also doesn’t qualify as UPopA. Onge and Australasians are equally out, since Mixe contain the least affinity to them among all Amerindians.
That still leaves two more potential candidates aside from Africans, namely
(a) La Brana-type WHG, which most likely isn’t sufficiently represented in modern French to create significants effects, and
(b) Afroasiatic groups, from Guanche/Berber to Arabs, which weren’t included in the “Eurasian” Panel.
2. Your argument is that D(Native American1, Native American2; Eurasian, Yoruba) shifts significantly towards Yoruba when Native American2 is Mixe. This doesn’t neccessarily mean that UPopA was an outgroup to all populations tested. It may also mean that UPopA had significant Yoruba-like (i.e. West African) admixture.
3. They “infer split off from Native Americans ~24.7 ka, ranging from 30-22 ka (95% CI).“. That is clearly after OOA, and also postdates the Ancient East Eurasian – West Eurasian split, making your “early OOA”, “early generic Eurasian” and “Archaic hominin” scenarios unlikely. Instead, a split 30-22 kya points towards UPopA coming from the Ancient West Eurasian line, most likely rather from the proto-WHG (Kostenki) than the ANE (Mal’ta) branch. That WHG-like population should have left little genetic traces in modern French, and the only candidate that comes to mind is La Brana,
4. While they assume a single admixture pulse, multiple pulses aren’t excluded, rather to the opposite (“the wide interval potentially reflects unmodeled continuous migration“). As such, we may well be dealing with two or more separate events, e.g. an early migration from Sub-Saharan Africa, plus a LBA Phoenician (Berber/ Guanche) migration.
The former is a/o indicated by aDNA analyses on the bottle gourd (Kistler e.a. 2014, https://doi.org/10.1073/pnas.1318678111) that demonstrated American bottle gourds (earliest attestations ca. 8 ka BC from Florida, ca. 7 ka BC from Oaxaca, Mexico) belonging to the African, not the Asian branch [both branches split ca. 140 kya]. American bottle gourds fall into various subclades – the a/m oldest samples e.g. cluster with Zimbabwe varieties, while a 300 BP sample from Baja California sits on the Zambian branch (TMRCA ca. 90 kya), which makes accidental introduction by ocean drift unlikely.
Phoenicians are a common speculation, e.g. for perceived similarities between Egyptian and Mesoamerican hieroglyphs and pyramids. In automated linguistic comparisons such as Greenhill e.a. 2010 (http://rspb.royalsocietypublishing.org/content/277/1693/2443), Mayan languages tend to associate with AfroAsiatic, especially Berber. But the Phoenician scenario will remain speculative, as long as we don’t have ancient Maya or Toltek aDNA for confirmation or rejection.
Whatever the origin(s) of UPopA – it is highly unlikely to have arrived in Mesoamerica from Asia. IMO, this is the first genetic evidence of transatlantic migration to the Americas prior to 400 BC .
[I hope this answers your question, Jacob.]
@FrankN
Yes, thanks for pointing out that Kennewick Man was already mtDNA X2a. That excludes any form of recent West Eurasian origin.
About the UPopA it’s really difficult to say without being able to test all the possible statistics. However, some comments on your remarks:
They systematically filter out French alleles in order to correct for a putative post-Columbus contamination (see Fig. 4B), thereby effectively depressing West Eurasian impact on the stats.
What I see in 4B is a simulation of filtering possible contamination (I think in LagoaSanta, being an ancient specimen, though I’m not totally sure I’m understanding that figure correctly). I don’t see anywhere that they systematically filter out French alleles in the statistics in Figure 4A.
That still leaves two more potential candidates aside from Africans, namely
(a) La Brana-type WHG, which most likely isn’t sufficiently represented in modern French to create significants effects
I think that any La Brana-type WHG admixture in Mixe would very clearly show up in any West Eurasian test population, especially in European ones. La Brana is a very strong ingroup for West Eurasians in general and Europeans in particular. (Same goes, but to a lesser degree, for Kostenki).
2. Your argument is that D(Native American1, Native American2; Eurasian, Yoruba) shifts significantly towards Yoruba when Native American2 is Mixe. This doesn’t neccessarily mean that UPopA was an outgroup to all populations tested. It may also mean that UPopA had significant Yoruba-like (i.e. West African) admixture.
In theory yes, but we’ve seen in Skoglund et al. 2016 (Ext. Data Table 2 & 3) that using Mbuti and Chimp as outgroups has the same effect as using Yoruba. So it doesn’t seem a Yoruba-specific thing, though indeed any SSA admixture in Mixe would make that stat positive, since they (SSA) are all outgroups to Eurasians.
3. They “infer split off from Native Americans ~24.7 ka, ranging from 30-22 ka (95% CI).“. That is clearly after OOA
Yes. But how reliable is that estimate of a split time of 24 kya when D-stats with all Eurasians are depressed by this mysterious admixture? It seems too strange that there was a Eurasian group c. 25 kya that is an outgroup to all known Eurasians (modern and ancient).
This leaves us with the possibility of North African (Berber-related) admixture that you mention. I guess this could depend on how much non-Eurasian related admixture such population could have. I would assume that if a stat like:
D(Mbuti, Sardinian; Mixe, Anzick1)
is positive (excess of allele sharing between Sardinian and Anzick1 when compared to Sardinian-Mixe) this rules out North African/Arabian admixture for groups where SSA admixture is not very high. It would be a matter of testing on one hand stats in the form:
D(Mbuti, Sardinian; NorthAfrican/Arabian, Anzick1)
to find the populations for which such stat is positive (Sardinian being closer to Anzick1 than to the NorthAfr/Arab pop). And then testing those populations like:
S(Mbuti, NorthAfrican/Arabian; Anzick1, Mixe)
to see if any of them is significantly positive. That would indeed support a transatlantic migration, but unfortunately I can’t test that myself.
BTW, the paper is also freely available here:
https://www.researchgate.net/publication/328815661_Early_human_dispersals_within_the_Americas
@ Alberto:
“I don’t see anywhere that they systematically filter out French alleles in the statistics in Figure 4A. ”
You are correct. It is probably worse than that.
The main paper states on p.2 “We merged these data with a SNP panel of 167 worldwide populations genotyped for 199,285 SNPs, enriched in NA populations whose European and African ancestry components have been identified and masked .” As concerns the Mixe data, reference is made to Reich e.a. 2012. In this respect, the SuppMat to Skoglund e.a. 2015 (https://media.nature.com/original/nature-assets/nature/journal/v525/n7567/extref/nature14895-s1.pdf) state: “The study that reported the Illumina array data [Footnote: Reich e.a. 2012] masked genomic segments of post-Columbian European or African ancestry.”
So, if I understand this correctly, all their analyses relate to Mixe data in which European and African ancestry components have been masked. If so, it is quite a miracle that they could identify UPopA at all, and it will be hopeless to try running any DStats in order to figure out one or several specific non-Asian/ non-Oceanic ancestry sources. Of course, this also supports your scepticism about their inferred split time.
Skoglund e.a. 2015 conducted some Chromopainter tests, described in SI 5, where they state: ” The Pima show an affinity to African Tshwa (Z = 3.71), possibly suggesting cryptic African ancestry. ” A statistically significant result, in spite of masked Sub-Saharan ancestry. Moreover, the Pima are not among the tribes well-known to have provided refuge to African slaves, and the Tshwa are not very likely to have been traded as slaves to the Americas. Interesting, isn’t it – even more so when considering that the Tshwa reside in Zimbabwe, which has supplied bottle gourd varieties to the Americas 10 kya or earlier.
Among the other affinities they noted, with a Z-score slightly below 3, are
– Mayan-Turkmen (Z=2.73)
– Kaqchikel-Ukrainians (Z=2.65)
– Quechua-Egyptians (Z=2.14)
Alright, Turkmens and to some extent also Ukrainians smell ANE, so this may be an artefact caused by one or more of the Trans-Pacific immigrations. But Quechua (Incas) tending Egyptian really got me thinking when I came across the Skoglund e.a. study back in 2015 …
@Frank
If that masking of genomic segments is true, then that’s rather disappointing. Any “cooked” genome should be labelled as such, so that it’s clear that we’re not dealing with the real ones.
I don’t even know which ones might be filtered and which ones not. With Global 25 I can see European admixture in several populations:
https://docs.google.com/spreadsheets/d/1P2Vk9M4Jh8Q0M99WnZiRWN5Oxv8wEmnTN-_NYcVKWHA/edit?usp=sharing
So it’s strange. Worse of all, it could be hiding real things or creating artefacts.
The linguistic diversity in the so called Amerind languages is very big. If you check the distribution of features on WALS https://wals.info/feature you will see that all variants are often present in America. Therefore, I would not be surprised if we found out that there were several pulses that contributed to the Amerind languages.
The highest amount of language isolates and the highest amount of diversity is on the North Pacific coast which is the area that a migration from Beringia first reaches:
(https://en.wikipedia.org/wiki/Indigenous_languages_of_the_Americas#/media/File:Langs_N.Amer.png)
YDNA diversity in America is not particularly big: Q-L54 (Q1a2a1), Q-M3 (Q1a2a1a1), and C-P39 (C2b1a1a). However, we know that yDNA is easily wiped out. If for example the amount of the Australasian in Surui is only c. 3 %, it it highly unlikely that there could be any yDNA left in them. We need a 20 kya old sample or older to see what this Australasian yDNA could be. The odds are quite high that it would be K2.
Maybe one day a skeleton that is old enough is accidentally found in a cave somewhere. Fortunately, it need not be in the Amazon, North America will probably do equally fine.
Frank, is there any way you could write this up concisely – it would surely shake the aDNA community up.
I’m certain of censorship of the Spirit Lake genome btw, we know this guy was a genuine redhead with light skin and wavy hair from his remains. You don’t even need to analyse the results they published – he was from the Si-Te-Cah. The truth is too big to cover up forever though, even if they succeed in North America where there are far fewer remains, there are thousands of remains in South America hidden away in private collections.
@Jacob:
The aDNA community has already been shaken up. I was surely not the only one to read through the Skoglund e.a. (D. Reich as co-author) 2015 SuppMaterials. And the first question is – why did they report those results at all, if they wanted to hide something. Answer – they didn’t want to hide facts, they wanted to shake up insiders.
I agree, there is a research bias, but that bias is coming from the North American archeological community that for decades was tied to the “Clovis first” paradigm that, btw, was never accepted in Latin America, especially not in Brazil (and note that this fresh study has three Brazilian co-authors). What aDNA research is currently focusing upon is taking that paradigm apart. Note the Skoglund e.a. 2015 title “Genetic evidence for two founding populations of the Americas”, followed up by Skoglund/ Reich 2016 describing 4-5 migrations (including Australasians, Na Dene, Paleoeskimos, and Inuit). In the paper discussed here, “Clovis first” isn’t even mentionned anymore. Instead, they describe the Clovis-WST dualism as established fact (which, IMO, is archeologically correct).
So, as I see it, “Clovis first” has now been successfully dismantled, and they move towards the next targets, which are (i) the Australasian signal and (ii) the apparently complex demographic history of the Pacific NW. One or two things at a time, that’s o.k. The Mixe stuff has been put on the agenda, and anybody reading their study carefully will arrive at similar conclusions as Alberto and myself. Have patience, give it some time. Sooner or later we will get aDNA from ancient Mayas, Toltecs, Aztecs, etc., and then we will all know more…
I don’t think there is any cover-up on Spirit Cave. In that case, they wouldn’t base demographic modelling on that sample. They could have stayed with Anzick1 for that purpose, as preceding studies have done, and refer in a footnote to Spirit Cave as some outlier, just as they did with BigBar. Their samples have been made publicly available and can be reviewed and analysed by everybody. Apparently, they haven’t yet checked on skin, eye and hair colour of the new samples, but someone will do that and either confirm or deny red-hairedness.
What I would nevertheless still like to see is a re-assessment of North Amerindian yDNA R1b1. The last thorough investigation that I am aware of was Bolnick e.a. 2008 (http://mbe.oxfordjournals.org/content/23/11/2161.full.pdf+html). They identified several mutations not shared with Europeans, some of which were 6-8 SNPs removed from the nearest European mutation (Fig. 6b). At the yFull calculation rate of 144 years per SNP mutation, this implies a split time of up to 1.2 kya, i.e. 850 CE, (somewhat) prior to documented Norse contact with North America. [Norse/ Icelanders are anyway not a likely source of R1b introgression – they should have introduced I1 in equal or even higher proportions, but I1 is comparatively rare among Native North Americans. R1b pre-dominance suggests Irelanders, Bretons, or Basques (whaling tradition!) as main source.] In the meantime, European R1b1 variants closer or identical to Amerindian ones may have shown up, reducing the European-Amerindian distance and inferred split time. But the Bolnick e.a. finding may as well still be valid to date. We cannot be sure as long as no re-assessment has taken place.
@Alberto: “I don’t even know which ones might be filtered and which ones not”
From what I have understood (but you may want to check it by yourself), the data from Reich e.a. 2012 has been filtered, while data generated by Skoglund e.a. 2015 (Apalai, Guaranai, Karitiana, Pima, Surui) remained unfiltered – they instead removed individuals with apparent European admix from their panel. The standard modern sets (Bolivians etc.) should be unfiltered. I am uncertain about the Mayan set – they carry some 10% R1b, and Skoglund e.a. 2015 report 17 of their 18 samples showing signs of post Colombian admixture, so some kind of filtering seems likely.
Some of your Global25 numbers, e.g. 1.7% Andalusian in Anzick1, and 1.1% Andalusian in Peru_3300BP, look pretty interesting. Before going further, may I however first suggest some changes to your model:
1. Chukchi are known to harbour Amerindian ancestry, and are as such problematic when trying to assess later ANE introgression. Skoglund e.a. 2015 have identified some ANE sources w/o significant Holocene Amerindian ancestry, e.g. Nganasan, Itelmen or Mansi. Might be better to use one of those instead. Alternatively, you may want to consult Sikora e.a. 2018 for adequate, unadmixed sources.
2. Koreans might approximate and conflate two potential admixture sources, namely Jomon (perceived parallels between Jomon and 4th mBC Ecuadorian pottery), and Chinese (shared rare gene mutations between the Chinese Crested Dog, the Aztec Xoloitzquintl, and the Peruvian Perro sin Pelo; NW American wolf DNA in traditional Chinese and Tibetan dog breeds). As such, replacing Koreans by (i) Ryukans (or Ainu), and (ii) Han could add information depth (or not, but that would also be an interesting find).
3. Adding Guanche to the model to check for possible NW African influence that in all likelyhood should have used the Canary Islands as stop-over when crossing the Atlantic.
4. Adding another South- Asian source. Skoglund e.a. 2015 from their Chromopainter analyses report Bolivian-Kinh (Vietnamese) affinity (Z=2.76). Atacama language displays manifold parallels (a/o numerals) to continental South-East Asia, in particular Tai-Kadai, but also Austronesian languages. Note in this context that the Coconut was introduced from the Phillipines to South America (Ecuador) around 250 BC (Baudoin/ Lebrun 2009, http://publications.cirad.fr/une_notice.php?dk=549869).
5. Finally, adding a SSA source couldn’t do harm. The bottle gourd apparently reached the Americas some 10 kya, and phylogeny points to a Southern African origin, so JuHoan might be an adequate proxy for the population involved. Alternatively, you could play around a bit with West Africans such as Yoruba (albeit I fear that the Yoruba – East Asian connection regularly popping up in Admixture graphs, possibly related to the Banana being attested in W. Africa from ca. 500 BC onwards, could introduce unwanted noise).
@FrankN
If Karitiana, Pima and Surui are unfiltered then that’s ok.
I just posted those models to show that post-Columbian admixture can be detected when it’s there. But anyway here’s another run with some of your suggestions (no Jomon/Ainu or Guanches available):
https://docs.google.com/spreadsheets/d/1tot615tunK7o0SEJ8066Gt90NBqhbBees-MMhpwao-I/edit?usp=sharing
These things should better be tested with formal methods anyway. I’ll look at the supplements in more details when I have time and might post an addendum if needed.
@Alberto
Thx for the new run! Some takeaways:
a) USR1 looks strange: 10.9% Onge, 5.4% Andalusian, 4.9% Han. The fit isn’t very good (distance = 5.6983), but still in an acceptable range for a sample of such high age, and your percentages are above noise level. I wonder what is going on there. Especially the Onge admix looks strange, as Moreno-Mayar 2018 report absence of the Australasian signal in USR1. The sample clearly deserves further analysis by formal methods.
b) Andalusian ancestry in modern populations (Quechua, Bolivians, Mixtec etc.) is probably reflecting post-Colombian admixture. Its absence from Mixe, but presence in Mixtec etc. demonstrates the a/m filtering applied to Mixe data.
3.6% Andalusian in Anzick1 and Kennewick looks strange. Some kind of artefact ? In that case probably the same artefact that also seems to affect USR1.
c) All Mesoamericans (Maya, Mixe, Mixtec, Pima) are shown with 2-3% Han admixture, which aligns with other evidence for Chinese-Mexican contact, e.g. from dog (a)DNA. There is even a written Chinese source reporting a monk’s visit to Mexico around 450 AD. I was initially sceptical about that source, but it seems to be genuine (see https://chapala.com/chapala/ojo2009/chinese.html for a detailed discussion of Chinese sources). The first European to have mentionned that source appears to be A. von Humboldt.
Here, I wonder why the Mesoamerican affinity to Chinese isn’t becoming apparent in Moreno-Mayar’s analyses.
d) No signs above noise level of Moroccan or Yoruba ancestry. I would still be interested about JuHoan as a source, but in general the available data doesn’t seem to support significant Transatlantic immigration.
@FrankN
Yes, USR1 is a very noisy sample. That’s why I removed it from the sources and placed in in the targets. I think it’s just a poor quality issue.
I was surprised too that the Spanish admixture in Anzick1 (Clovis) and Kennewick didn’t go away when including Yoruba. I thought it was DNA damage that showed up as Spanish for lack of a more basal source, and expected Yoruba to take it, but it didn’t. So now I’ve included English in addition to Spanish to see if it’s rather contamination. And so it seems, now both samples showing ~4% English (second tab, same link above).
I changed Yoruba for Ju Hoan North, but no difference there.
Alberto – I see, and English improve the fit slightly. One possibility may be that the model tries to trim down the Australasian signal and uses West Europeans for that purpose. Maybe you include MA1 and see if the English signal persists.
Otherwise: Several earlier analyses of American dog aDNA suggested an introduction from Europe. Thalmann e.a. 2013 (https://www.researchgate.net/publication/258529165_Complete_Mitochondrial_Genomes_of_Ancient_Canids_Suggest_a_European_Origin_of_Domestic_Dogs), e.g., demonstrated that a 10,000 BP dog from Illinois (recently re-dated), as well as several later, but still pre-contact North American dogs, formed a clade with a 14,500 BP wolf-like dog from Switzerland.
Leathlobhair e.a. 2018 (https://www.researchgate.net/publication/326211519_The_evolutionary_history_of_dogs_in_the_Americas) tell a somewhat different story – they connect Paleoindian dogs to recently discovered dogs from Zhokhov Island, off the New Siberian Islands in the Arctic Ocean. That is somewhat anachronistic – the earliest attestations of domestic dogs in N. America (Iowa/ Illinois) are from ca, 10,000 BP, a millenium earlier than the Zhokhov dogs. However, older sites in Central North Siberia may now be flooded by the Arctic Ocean.
It becomes more and more apparent that the earliest Paleoindians didn’t have dogs, and dogs reached the Pacific coast comparatively late. On the Californian Channel Islands, possibly the archeologically best documented area in the USA, human settlement is evidenced at least since 13,000 BP, but dogs only appear around 6,000 BP, four millenia later than on the Upper Missisippi. This suggests an early holocene immigration from Central North Siberia, apparently along the Arctic Ocean and then up the Mackenzie that at that time served as outlet of the large Agassiz meltwater lake and the Great Lakes basin.
While Leathlobhair e.a. 2018 didn’t find a connection between West Eurasian and ancient Siberian/ Paleoindian dogs (but also didn’t put much effort in such analysis, e.g. by including West Eurasian dog aDNA in their modelling), other studies have arrived at different conclusions. Wang e.a. 2016 (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4816135/) state: “Around 15 000 years ago, a subset of [South Chinese] ancestral dogs started migrating to the Middle East, Africa and Europe (..) One of the out of Asia lineages also migrated back to the east, creating a series of admixed populations with the endemic Asian lineages in northern China before migrating to the New World.”
Assuming that those migrating dogs were accompanied by humans, those Central North Siberians bringing dogs to North America may well have incorporated a bit of West Eurasian ancestry. In Wang e.a., Amerindian breeds cluster with the Belgian Malinois, while Thalmann e.a. 2013 found connection to a Swiss wolf. Against this background, it might be worthwhile to try either Bichon or Loschbour in your model.
@FrankN
Yes, that’s right. I dropped AfontovaGora3 from the last run too without even noticing. Now adding it again brings things back to normal (only noise levels in Anzick and Kennewick of contamination). It’s in the 3rd tab from the same link above. (I tried with MA1 too, but all took AG3 instead. Loschbour didn’t change things either).
More importantly, though, I found out where these ancient samples included here come from:
https://www.cell.com/cell/fulltext/S0092-8674(18)31380-1
As usual, two teams published two papers about the same subject at the same time. And I missed this last one from the Reich lab team. I’ll update the post about it if I have time.
@Alberto:
That Posth e.a. study is also great, thx. for the link. If you decide to update the post, I suggest to include both their Migration map (Graphical Abstract) and the PCA (Fig. S2D).
The PCA shows that Native Americans distribute along three clines, which more or less intersect between the Wayuu (Arawak, on the Columbian and Venezuelan Caribic coast), and Mixe/ Pima.
– One cline starts in the upper left corner with the Surui. I suppose that cline relates to the Australasian (& Denisovan) signal. Intriguingly, however, their admixture Graphs (Fig S5 I-G) model Surui not as Onge, but as East-Asian admixed (7-8%). Moreover, according to them the respective admixture wasn’t present yet in the ancient Peruvian, Chilean and Brazilian samples investigated, which is somewhat at odds with your nMonte results and previous research on that Australasian signal.
– The second cline starts in the lower left corner with Costa Rican Cabecar, runs through other populations from Costa Rica and Panama such as Teribe, Embera and Guaymi. This cline should reflect the proper “Mesoamerican” / UPopA signal that apparently is already watered down in Mixe, and seems to have originated on the Isthmus of Panama rather than in Mexico. Interestingly, the Belize_9300 BP sample isn’t located on that cline, suggesting the signal wasn’t yet widespread across Mesoamerica by that time.
– The third cline runs relatively horizontal. Its endpoint to the right is marked by Nganasan. Obviously Paleosiberians, ANE/ENA, and later trans-Beringian migrations.
The most “Siberian” of the ancient samples are USR1 and historic Athabascans (100 BP). Kennewick, 939 and Lucier (S. Ontario) are also visibly pulled towards ENA. That would be the NNA population identified in Moreno-Mayar 2018, albeit I wonder whether they represent an original founding population, or a later (Younger Dryas/ Early Holocene) migration wave from Central Siberia. Archeology suggests the latter – around 8,000 BC there was a clear cultural break in the American Midwest (the post-Clovis to Archaic transition) that, aside from the presence of dogs, a/o also included increased sedentarism, stronger use of plant ressources, e.g. nuts, a.o.m. Your nMonte results, showing that AG3 works as additional source, but MA1 not, seem to support the idea of a later immigration that incorporated Central Siberian elements.
Intriguingly, their admixture graphs consistently show 2% non-Asian admixture in Lucier (S. Ontario, 4,800-500 BP) “that likely reflects a low level of contamination in these samples“. Of course – these stupid Canadians can’t treat their samples properly, But wait – there is no such contamination found in the samples from the Pacific NW…
Another interesting note is that Anzick1 clusters with mid-Holocene Channel Island inhabitants and historical Chumash speakers (one more puzzle piece for better understanding the genesis of linguistic diversity in the Pacific NW). As Clovis culture never reached the Channel Islands, which instead have yielded evidence of WST occupation (https://insider.si.edu/2011/03/californias-channel-islands-may-have-once-held-north-americas-earliest-seafaring-economy/) this observation makes me even more sceptical wether Anzick1 really represents a member of the Clovis culture. Similarly, early Brazilian sites such as Lapa do Santo, and early Argentinian sites, while genetically related to Anzick1, produced arrowheads much different from Clovis points (http://revistapesquisa.fapesp.br/en/2018/06/25/occupation-of-primordial-brazil/).
Note following on from this paper and the Yana paper (Sikora et al), this new paper Pinotti 2018 (https://www.cell.com/current-biology/fulltext/S0960-9822(18)31495-7) provides further support for the “short Beringian standstill” model*.
Major conclusions:
“- A Beringian Standstill of <4,600 years led to both Siberian and American Y-lineages" (specifically: "we estimate a Beringian Standstill duration of 2.7 ky or 4.6 ky")
"- Y-lineage split times rule out occupation of the Americas before 19,500 years ago"
More evidence that fits with the Sikora paper's model that "Between 20 and 11 kya, the ANS population was largely replaced by peoples with ancestry from East Asia, giving rise to ancestral Native Americans and “Ancient Paleosiberians” (AP), represented by a 9.8 kya skeleton from Kolyma River. ".
Particularly the short date 2.7kya would be easy to reconcile with a chronology where the glacial maximum peak is about 26kya, then deglaciation begins 20kya and for a fairly short time before this, populations expand out of East Asia into the Beringian refugium (mixing with ANS Siberian populations to some degree), then finally entering the Americas as a ice free routes come about around 16-15kya.
(I don't know how well this would fit with any models of deep splits within Native American populations about 30-22kya as seems to be in quoted model for the paper for this post. That seems not to work. Diversification and population structure within Native Americans at 17kya-14kya seems to fit better.).
*As opposed to the long standstill of 15ky, which was based on speculation that Yana samples were genomically the same as population as later Beringian populations, which we know to be wrong.
@Matt
Thanks for the update. I think that paper makes good sense and confirms what we more or less know.
I didn’t update the post after reading the Posth, Nakatsuka et al. paper more carefully because I became rather sceptic about several points raised in these papers (and being a subject beyond the scope of this blog I didn’t want to discuss in detail about it). But for the record:
My impression is that they’re reading a bit too much into too little. Looking a the detailed statistics there’s really not much there that is consistent enough to draw the kind of conclusions they do. Maybe the team from this second paper is a bit too attached to the idea of population replacements as a rule (rather than an exception), or just not seeing the bigger picture and focusing on a few stats that stand out.
Overall, I think that these papers are very interesting for the history of the Americas, and they show that it’s a bit more complicated than a single pulse of migration as the founding one of all Native Americans. The evidence for an ongoing gene flow (maybe intermittent) looks more or less clear. Anzick-1 might already harbour some of this post Main Pulse admixture, and it becomes very evident in Kennewick Man. Probably this explains at least partially the dilution of the Australasian signal in North American populations. The rest I wonder if it could be due to selection for alleles for warm climate adaptation that where selected against in NE Asia and then again selected for in South America (counter intuitively bringing a population closer to an ancestral one by drift/selection).
For the “Unknown Population A” I’d like to see some evidence in ancient samples before I trust that it’s not an artefact of either genome manipulation in the Mixe samples or some cryptic post-Columbian admixture that involved a low amount of African one.
Presuming that the paleo-American signal is true and testifies to an earlier entry to the Americas, the new paper by Pinotti et al is interesting as it shows that, while Q lines and C-P39 are estimated to be c. 15 kya old:
”the South American C3∗, henceforth named C3-MBP373, is the first branch to diverge within C3-L1373 from the other, mainly Siberian clade C3-F3914, at 21.6 (19.1–24.4) kya.”
(https://www.cell.com/current-biology/fulltext/S0960-9822(18)31495-7)
The new mtDNA paper by Llamas et al gives new age estimates to the main Native American mtDNA lineages (fig. 3):
D1 age in America 18,5 ka, separation from Asian lines 27,5 ka
C1 age in America 18 ka, separation from Asian lines 20 ka
B2 age in America 17 ka, separation from Asian lines 24 ka
A2 age in America 16,5 ka, separation from Asian lines 22,5 ka
(http://advances.sciencemag.org/content/2/4/e1501385?fbclid=IwAR2Yt4KjdFa6SToVuKb1tBc1hZhqNrpB-T5-TLYnTnpFv0ZTm4vVW8DD8ro)
Among the above mtDNA lineages, D1 is the oldest and separated from the Asian lineages clearly earlier than other lineages.
Interestingly, there is another paper by de Saint Pierre in which it was stated that ”the mitochondrial subhaplogroup D1g described in 2012 and found in Amerindian populations of southern Chile and Argentina, represents today as Monte Verde did before, an interesting paradox that has not yet received much attention. The age calculated for D1g, between 25,000 and 19,000 cal yr BP is extremely old for a South American mitochondrial subhaplogroup. The anomalous age of this haplogroup does not fit the currently accepted framework for the other mtDNA haplogroups in the Americas.” (https://www.sciencedirect.com/science/article/pii/S1040618216310941)
D1 was also detected in a c. 12,000 years old female with paleo-American phenotype found in a cave in Mexico (http://science.sciencemag.org/content/344/6185/750)
Therefore, it is possible that there was an earlier entry to Americas of yDNA C-MBP373 and mtDNA D1 with paleo-American phenotype but these first Americans were severely pruned by more successful later migrations.
The ancient yDNA has shown us that in particular y lines become constantly extinct. Therefore, the age of surviving y lines does not tell the whole story as there has been a huge amount of extinct y lines in the Paleolithic and Mesolithic populations and we cannot rule out the possibility that there were such extinct y line(s) in America.
The date range for unpopA is interesting: “11% gene flow from UPopA into Mixe ~8.7 ka (95% CI: 0.4-13.9 ka; the wide interval potentially reflects unmodeled continuous migration)” This upper end of that time frame coincides with the onset of the younger dryas. It also coincides with a shift in population centers in the nevada/Texas regions. Prior to about 13K, you had two populations the Western Stemmed population in Nevada and the Clovis population in Texas. After this you see the population centers merge: https://www.pnas.org/content/112/39/12127 (look at the supplemental for more detail chronology).
It is really odd that these papers look really hard at contemporary Eurasian populations, but do not really dig into the ancient DNA Eurasian samples. Should the X2 genome in NA come from non-beringia populations, it obviously happened a long time ago. Based on Kennewick and age of the various X2a and X2g haplogroups.
The papers around X2 are really interesting.
Timing and geography make a Beringia route for the X2 mtDNA haplotypes into the Americas problematic. Further, no ancient or contemporary DNA samples have been analyzed that indicate a clear migration history of the X2 mtDNA haplogroups across Asia and into the Americas from Beringia (Oppenheimer et al. 2014). However, three separate papers state that the X2 haplotype entered into North America from Beringia (Hooshiar Kashani et al. 2012, Fagundes et al. 2008, Perego et al. 2009). These papers all rely on the same central argument, temporal concordance of X2a mtDNA haplogroup coalescences timing to other Native American mtDNA haplotypes. Fagundes et al. (2008) examines a broad swath of mtDNA which only contains two X2a mtDNA and does not include the rare X2g type found by Perego et al. (2009). Kashani et al. (2014) examines no new X2a mtDNA haplotype data but uses correlation to another Native American haplotype to support the model. Further, statements within the Kashani et al. (2014) paper directly conflict as they state: “…a temporally distinct arrival for C4c cannot be completely dismissed, the similarities in both ages and geographical distributions between C4c and X2a suggest that these two lineages possibly arrived together from Beringia [emphasis added]” but then go on to claim that “the finding that C4c and X2a are characterized by parallel genetic histories definitively dismisses the controversial hypothesis of an Atlantic glacial entry route into North America [emphasis added].” In addition, age correlation between C4c and X2a mtDNA haplogroups is weak as they do not consider the presence of X2g which suggests that X2 haplogroups could have entered the Americas near the time the two X2 mtDNA haplogroups diverged. Further, the geographical alignment of C and X2 mtDNA hapolgroups is weak as the haplogroups originate in either East Asia or in the Near East, respectively, and do not have a similarity in distribution or geographical range until the arrival of C4c mtDNA haplogroups into the Americas around 12,000 years ago (Derenko et al. 2010, Hooshiar Kashani et al. 2012). Overall, these data are insufficient to assert that the Beringia route is the path taken for the distribution of X2 mtDNA haplogroup.
What is interesting is that C4 mtDNA haplogroups are found in Western Eurasia in the ancient DNA samples. What would be interesting is if C4 in the America’s is NOT a sign of Beringia populations but of some paleolithic western eurasian population-with potential migration across the North american continent and back into asia via Beringia
Raff and Bolnick (2015) suggest that the Kennewick man data supports a Beringia route for X2a dissemination into the Americas as it: “places the oldest and most basal X2a lineage in the Pacific Northwest” (Raff and Bolnick 2015). This assertion ignores the fact that Kennewick man dates to nearly 4,000 years following even conservative estimates for entry into the Americas. In contrast, the data does suggest that Kennewick man could be representative of a maternally ancestral population to the Yakima people and the Nuu-Chah-Nuluth people (Brown et al. 1998). Both these people carry a X2a mtDNA haplogroup that does not appear to be X2a2 or X2a1 present in the Ojibwa or other North American Natives and likely represent the autochthonous development of a unique X2a mtDNA haplogroup (Perego et al. 2009). As it is known that a population with the Clovis technology made it to the west coast, more specifically found in East Wenatchee Washington (Wilke et al. 1991), it is possible that an eastern North American population migrated to the Pacific Northwest America and admixed with the populations from Beringia (Collins et al. 2013). Regardless, an east to west migration model is consistent with both main models for entry of the X2 mtDNA haplogroups into the Americas.
X2 mtDNA haplogroups tend to be rarely found in most populations, ancient evidence could be difficult to find and could, theoretically, be lost in contemporary populations due to extinction of the lineage. HOWEVER, the X2 haplogroups developed ~22,000 years ago in the Near East, migration into the Americas is temporally and geographically problematic. To have occurred, a population carrying the X2 haplogroup would have had to cross Central Asia and Siberia during the LGM and arrived in Beringia an estimated ~1,000 years prior to the estimated dates for coalescence of the X2 haplogroup. Overall, the presently available genetic data does not support the Beringia hypothesis suggesting that other routes for X2 mtDNA haplogroups into the Americas need to be considered.