There’s a new amazing preprint at bioRxiv about the population history of Siberia, which also deals with the origin of Native Americans. The new study brings a good amount of new samples spanning from the UP (~32 kya) to ca. 1250 AD that clarify the genetic history of the region. While it does not show anything too surprising or new, it basically confirms with samples and provides more accurate dates and places for what could already be envisaged from the previous limited amount of aDNA available. Here’s the abstract (emphasis mine):
Far northeastern Siberia has been occupied by humans for more than 40 thousand years. Yet, owing to a scarcity of early archaeological sites and human remains, its population history and relationship to ancient and modern populations across Eurasia and the Americas are poorly understood. Here, we report 34 ancient genome sequences, including two from fragmented milk teeth found at the ~31.6 thousand-year-old (kya) Yana RHS site, the earliest and northernmost Pleistocene human remains found. These genomes reveal complex patterns of past population admixture and replacement events throughout northeastern Siberia, with evidence for at least three large-scale human migrations into the region. The first inhabitants, a previously unknown population of “Ancient North Siberians” (ANS), represented by Yana RHS, diverged ~38 kya from Western Eurasians, soon after the latter split from East Asians. Between 20 and 11 kya, the ANS population was largely replaced by peoples with ancestry from East Asia, giving rise to ancestral Native Americans and “Ancient Paleosiberians” (AP), represented by a 9.8 kya skeleton from Kolyma River. AP are closely related to the Siberian ancestors of Native Americans, and ancestral to contemporary communities such as Koryaks and Itelmen. Paleoclimatic modelling shows evidence for a refuge during the last glacial maximum (LGM) in southeastern Beringia, suggesting Beringia as a possible location for the admixture forming both ancestral Native Americans and AP. Between 11 and 4 kya, AP were in turn largely replaced by another group of peoples with ancestry from East Asia, the “Neosiberians” from which many contemporary Siberians derive. We detect additional gene flow events in both directions across the Bering Strait during this time, influencing the genetic composition of Inuit, as well as Na Dene-speaking Northern Native Americans, whose Siberian-related ancestry components is closely related to AP. Our analyses reveal that the population history of northeastern Siberia was highly dynamic, starting in the Late Pleistocene and continuing well into the Late Holocene. The pattern observed in northeastern Siberia, with earlier, once widespread populations being replaced by distinct peoples, seems to have taken place across northern Eurasia, as far west as Scandinavia.
So let’s start with the oldest samples from Yana RHS, dating to around 31.6 kya and referred to as Ancient North Siberians (ANS). This represent a new population in the ancient DNA (aDNA) record, at least partially so. A note a bout the name: whenever a new population is found it needs a name, which usually corresponds to the geographical location where the specific samples are found, and sometimes to the economical subsistence strategy or the time when they lived. I this case, the choice of Ancient North Siberian does not imply that this population was restricted at that time to North Siberia (since it’s very likely that people living in South Siberia at that time were part of the same population), nor does it imply the place where it originated (since it’s very unlikely that it was actually in North Siberia). So let’s just take the name for what it is: a way to refer to this population due to the location and time of the samples.
So who were these ANS? Probably many already have guessed that they must be related to another population known as the Ancient North Eurasians (ANE). Indeed, ANS represent an early stage in the development of the ANE population, and therefor ancestral to ANE, as seen both in the autosomes and the Y chromosome (P1 in these 2 samples). This population is somewhat intermediate between West Eurasians (WE) and East Eurasians (EE), but closer to West Eurasians (it can be modelled as 75% WE and 25% EE). Whether this is because the split from West Eurasians soon after the split between WE and EE alone or because of subsequent admixture with EE is unclear, though in theory I guess that admixture is the only way to explain a higher affinity to EE (or otherwise WE admixed into some other more divergent ghost population that separated them further from EE, but this is quite more speculative).
As for the origin of this ANS population, it seems quite clear that it was to the south west. A population that is in between WE and EE must be from somewhere around Central Eurasia. And after all, ANE was already in high proportions in 10 kya samples from West Iran, in pre-BA sample presumably from the Indus Valley Civilization and it’s present all the way to South Indian tribals.
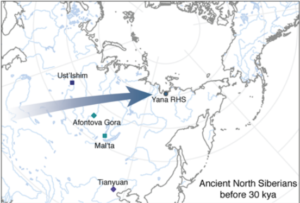
The paper draws what I think it’s a very good picture of the dynamics of the populations that occupied difficult climatic zones such as Siberia during the last glacial period. While it’s amazing that populations could actually live in North Siberia at that time, it’s not surprising that whatever subsistence strategies and ecological niches they managed to find and exploit, they were quite fragile and could easily break with any change in the climatic conditions. The paper documents the discontinuities during, for example, the LGM and investigates through climate modelling the possible areas where these populations might have found refuge during each period.
Following the LGM, this ANS population is replaced by a different one, the Ancient Paleosiberians (AP), who are the ones who went further to colonize the American continent. This population was partly descended from ANS, via ANE, but had a large amount of East Asian admixture (from around North China/SE Siberia, represented by other ancient samples from the Devil’s Gate cave). In this part is where I find the only issue with an otherwise exemplary paper. When arguing the possible place where this AP population formed, they write the following:
Conditions in the region became less suitable during the LGM, supporting the lack of genetic continuity between Yana RHS and later groups. Interestingly, we find evidence for a potential refuge in southeastern Beringia, as well as a possible coastal corridor along the Sea of Okhotsk and the Kamchatka Peninsula during the LGM (e.g. panel 22ka, Alaska, Extended Data Fig. 8a), in line with previous reports48. A possible scenario for the ANS gene flow during the formation of Native Americans and AP might therefore have involved early ANS surviving in southeastern Beringia during the LGM, with subsequent admixture with EEA [Early East Asians] arriving by a coastal migration ~20 kya. This scenario would also be consistent with a divergence of Ancient Beringians from ancestral Native Americans in eastern Beringia rather than in Siberia, which is also supported by genetic data (Scenario 2 in41).
I think that this scenario is incompatible with their current genetic data. The “ANS gene flow during the formation of Native Americans” is not a precise statement, since it’s not really ANS, but more precisely ANE. Their scenario implies that the evolution from ANS to ANE happened simultaneously and independently in Beringia/Kamchatka and in South Siberia (Laka Baikal) and SW Siberia (Altai), something that is not really possible. They show in different stats and figures in the paper that the Malta1 (near Lake Baikal, ca. 24 kya) shares more drift with Native Americans than the Yana samples. And AfontovaGora3 (Altai region, 17 kya) shares even more drift with Native Americans than Malta1 does. This makes is basically impossible that ANS survived in NE Siberia and contributed to the formation of Native Americans (it would require a very convoluted scenario of back migrations from NE Siberia to Altai that I won’t extend in outlining here due to its various complications).
So what it seems to have really happened is that those ANS populations from N/NE Siberia went extinct sooner or later, and subsequently the area was repopulated from people coming from around the Amur basin who mixed with people coming from much further west, maybe the Altai region. This seems rather clear, as the evolution from ANS to ANE seems to imply an increased affinity to West Eurasians (shown in some graphs as admixture from a branch related to CHG, but without Basal Eurasian admixture). They go on saying:
Alternatively, the closer affinity of both Kolyma1 and Native Americans to Mal’ta rather than Yana could suggest a more southern location (Lake Baikal region) for the admixture, with a subsequent northward expansion following the LGM. While the archaeological evidence of a movement south during the LGM supports this scenario, the genetic isolation between Asians and ancestral Native Americans after ~24 kya would have been difficult to maintain if several populations sought refuge in the region. Our results nevertheless support the broader implication that glacial and post-glacial climate change was a major driver of human population history across Northern Eurasia.
I’m not really sure what they mean by the “genetic isolation between Asians and ancestral Native Americans after ~24 kya” (I might have missed the specific part in the paper – will check), but in any case this could mean that it was these EEA who were in the Beringia and/or Kamchatka area prior to the LGM, since we don’t have direct genetic evidence from that region at that time. Or any other scenario. But clearly (IMHO) not the first one proposed.
Regarding the higher affinity of ANE to some WE branch related to CHG (I wouldn’t take this very literally, since it probably means something more similar to Kostenki), it further goes on to show that in the formation of EHG there is admixture fro CHG (this time CHG-proper, or closely related), and shows the presence of Basal Eurasian admixture in EHG too. This has been known by many of us for a long time now, but it’s the first time it appears documented in a scientific paper, so that’s another good point in the paper.
Continuing with Native Americans, there’s this other mysterious Australasian (Onge/Papuan related) admixture found in previous studies in populations from the Amazon, like Surui and Karitiana, specifically when compared to the Mixe and Pima populations from the SW part of North America. For what I see here, however, the relatedness of the Yana samples to Anzick-1 (a 12.4 kya sample from the Clovis complex) and to Karitiana and Surui is symmetrical, while it becomes asymmetrical with Mixe and Pima. This suggests that whatever Australasian “admixture” (if it can be called admixture at all) present in modern samples from the Amazon might have been already present in the Anzick-1 sample, and it’s rather Mixe and Pima who would have later experienced some genetic shift that makes them further away from Australasians (but apparently IIRC, there’s no evidence of admixture from any known source in Pima and Mixe, which makes the case complicated. Maybe if these 2 population went through a bottleneck that triggered a strong selection that affected specific regions of the genome associated with Australasians it could lead to these results, but this is something that’s purely speculative on my part and would need to be tested as an alternative).
Holocene transformations across Siberia and Beringia
The paper further investigates the Holocene genetic shifts across Siberia, but I’m afraid that in order to keep this post at a reasonable length I’ll have to just outline a few headlines and leave further analysis for either a second part or the comments section.
- A second migration from Beringia to North America and Greenland represented by the ancient Saqqaq sample (Paleoeskimo, from Greenland) harbouring an excess of DGC-related ancestry.
- A back migration of Native Americans to Siberia represented by Uelen and Ekven Neo-Eskimo sites (2.7-1.6 kya) that strongly resemble contemporary Inuit and could harbour some 30% Native American admixture
- The replacement of Ancient Paleosiberians by Neosiberians in most of Siberia. While they show that AP ancestry was still common by the Early Bronze Age, by the late Bronze Age it was largely restricted to NE Siberia, with populations that closely resemble contemporary Koryak and Itelmen.

Finally, there’s a linguistic discussion on the supplements that I think it’s quite an interesting read. I find that they have managed to get a good understanding of the genetic and archaeological data to try to correlate it with the linguistic hypothesis that exist so far. It’s an informed and balanced summary that doesn’t take any strong stance (which would be quite difficult to do knowing the region we’re dealing with). I think that maybe some of the references to Uralic or to macrofamilies might be already a bit outdated, but that’s not a big issue since they’re just mentioning what’s been hypothesized so far, and anyway it’s not the place or time to go into such issue. Overall, I think that it’s a necessary addition to any aDNA paper, much in line with what I wrote about in a recent post about it.
[UPDATE 27th June 2019: the final version has been published already with an important correction about the issue discussed above:For both Ancient Palaeo-Siberians and Native Americans, ANS-related ancestry is more closely related to Mal’ta than to the Yana individuals (Extended Data Fig. 3f), which rejects the hypothesis that the Yana lineage contributed directly to later Ancient Palaeo-Siberians or Native American groups.
Good news to see that part being addressed in the final version.]

As to the origin of the ANE population, I think Lipson et al. (2017) have shown that the East Eurasian component in ANE was most closely related to present day Ami. The models I’ve played around with indicate that Indonesians and Malays provide an even better fit, but it’s not something I’ve explored in depth. The source seems broadly speaking ‘Austronesian’ in any case. I think it probably isn’t a coincidence therefore that the phylogeography of Y-DNA K2 points to a spatially relatively confined diversification in Southeast Asia.
@Marko
That’s interesting. I haven’t looked myself into this because I lack the tools for it. I seem to recall that in the case of MA1 it would be more related to Onge then to Ami, but in AfontovaGora3 I don’t know if this holds true.
I think that being somewhere in Central Eurasia makes more sense than being in Eastern Eurasia, given the affinities of ANE. So the ENA part must have been somewhere closer to Central Eurasia too, I guess.
@Alberto
Definitely, those components must have met somewhere half-way geographically. I would assume that older samples from North India and the Himalayas would be informative in that respect.
I also believe that Iran_Neo derives from an ANE-like population that was more ENA shifted than the Siberian samples, which would fit nicely if the components met in Central Asia – Sungir from the north-west, ENA from the south-east. This encounter likely gave rise to hybrid populations with differing proportions of Sungir/ENA.
I agree with you when you argue against the idea of a survival of the ANS population in Beringia and their admixture with East Asians only there. You presented counter-arguments based on the autosomal data, but I would like to add that also the uniparentals are against that scenario. ANS are mtDNA U2’3’4’7’8’9, but that mtDNA is non existant in Native Americans. MtDNA usually survives quite well compared to yDNA, and I do not think that it is plausible that all ANE mtDNAs would have been lost. Moreover, yDNA of ANS samples is not Q but P1. They are on a different branch from R and Q. It is interesting that TMRCA of P on yfull is c. 32 kya which is close to the age of Yana samples. I would think that this is the time frame when P starts spreading to the north from Central Asia. TMRCA of Q is c. 29 kya, i.e. c. 3 kya younger than the Yana samples. I presume that the Q population that gave rise to Native Americans arose in another location. The area of expansion of P must have been very vast as it is today found in Island South East Asia. Therefore, there is no reason to presume that Q arose in the same location as P1 of the Yana population.
“It would require a very convoluted scenario of back migrations from NE Siberia to Altai that I won’t extend in outlining here due to its various complications.”
What complications? Hunter-gatherers that far north wouldn’t just sit and wait until they’re frozen to death when the LGM kicked in. They were there for a reason: Hunting mammoths and they would follow the mammoth herds. So they went where the mammoths went and the mammoths probably went back and forth. So yes, I can imagine a group of mammoth hunter around Lake Baikal 24 kya to be descendant from a group of mammoth hunters from around Yana river.
http://rspb.royalsocietypublishing.org/content/280/1770/20131910
To be short: Hunter-gatherers don’t really migrate like farmers or pastoral tribes. They follow herds.
@ Alberto
”I think that this scenario is incompatible with their current genetic data. The “ANS gene flow during the formation of Native Americans” is not a precise statement, since it’s not really ANS, but more precisely ANE. Their scenario implies that the evolution from ANS to ANE happened simultaneously and independently in Beringia/Kamchatka and in South Siberia (Laka Baikal) and SW Siberia (Altai), something that is not really possible. ….
So what it seems to have really happened is that those ANS populations from N/NE Siberia went extinct sooner or later, and subsequently the area was repopulated from people coming from around the Amur basin who mixed with people coming from much further west, maybe the Altai region. …”
Agreed. They seem to base their opinion of paleoclimatological modelling, which is very interesting, but ultimately a more solid scenario in keeping with archaeology and genetics is as you have outlined.
Just another minor point to an otherwise excellent paper – they might have wished to comment more on Ust-Ishm, the 45 ky old Initial Paleolithic individual from West Siberia. Populations like Yana and MA-1 must represent discontinuities at least on a regional level.
So as amazing as it is that humans reached the arctic by 30 ky BP, it seems this colonization was tentative, and continuous settlement of the arctic was only achieved much later on (after the LGM).
Instead, the initial peopling of Siberia, as Europe took at least a couple of attempts. I am curious where the starting point, or ”base-camp ”, might be for this ? I recently learned that one possibility is the Zagros region. Intuitively, this might seem far too south, however, that is where the Paleoarctic and the Savanah-Arabian ecotones (broady speaking) intersected back 40 k years ago. (”Kaldar cave”..Bazgir et al).
Its very likely at this point that these ANE populations where responsible for spreading R1a Y-dna into eastern europe by mixing with the women there, giving rise to modern eastern europeans.
@Robert & Kristiina
Thanks for adding further about that part of the paper I commented about. It seems strange to me that while the paper has that genetic data in it and everything else is well explained and understood, they still went for that scenario of survival in NE Siberia. It’s still a preprint, so maybe in a revision they will consider these comments about it.
@Robert & Marko
Yes, it would be great to have some aDNA from the Zagros to SC Asia from that period in the UP. It would probably help to explain a lot about the population dynamics in more northern parts of Eurasia. I’ll check that reference about Bazgir et al. thanks.
@epoch
Though that scenario still wouldn’t support the mentioned survival in Beringia/Kamchatka? In order for this latter scenario to be true it requires a more complicated series of events, so that AfontovaGora3 and Native Americans descend from the same ANE population that survived the LGM in that area in NE Siberia.
@Robert and everyone interested
Re: the basecamp question, there is an interesting and short overview of Central Asian sites with dates in Marcel Otte’s paper ‘Central Asia as a Core Area’ (2014). I think the sites mentioned there ultimately hold the key to understanding West Eurasian genetic variation.
Moving further down in time- another interesting discussion point about Siberia is what happens during the LGM. One school of thought sees Siberia to have been continuously occupied (Kuzmin & Keates, eg) whilst others see depopulation to archaeologically impoerceptible levels (eg Graf, Goebbels). The debate revolves around the speifics and validity of certain C14 dates around the LGM.
Genetics suggests that MA-1 (c. 25,000 cal BP) is from a similar population to Afontova-Gora individuals (although of poorer coverage) which appear c. 18-16,000 cal BP. However, this does not exclude the possibility that there was a depopuation for a couple of thousand years, followed by a re-settlement by similar groups. Indeed, these new groups bore a new material culture, new techniques fo hunting, etc. Graf specifically points to Hakkaido (Japan) as a possible place of origin for the microblade technique. However, the said similarity between AG-3 and MA-1 excludes this scenario in purity.
An article Frank suggetsed a while ago (The fundamental hominin niche in late Pleistocene Central Asia: a preliminary refugium mode – Beeton et al) points to the possibilty of the Tien Shan mountains as a refuge. Here they would have had contacts with microblade -bearing groups from Mongolia & further east, then spread back north to Siberia.
Hopefully some more LUP data from Siberia and central Asia will help.
I forget the gazette, but i recall it was reported that 2 LUP individuals ”cooking” in a couple of Labs (one was from Turkmenistan, ..)
@ Marko
Thanks, I am familiar with Otte’s work. I think it is making sense in light of the emerging data.
Alberto:
I don’t think they say that Ancient Paleosibirians and ancestral Native Americans are the same population, which would be quite a stupid statement to make considering that their AP sample is just 10,000 years old, while Beringian colonisation of the Americas possibly already commenced some 18-20 kya. If you read their abstract again, you will see that they instead point towards the same process, namely the expansion of East Asian ancestry, underlying the formation of both ancestries. And, in fact, their admixture graphs (Fig. 2, S6.11) indicate the following (DC= Devil’s Cave):
– AP ~ 75% DC, 25% MA1
– AP ~ 67% DC, 33% Yana
– Anc. NA ~ 63% DC, 37% MA1
– Anc. NA ~ 59% DC, 41% Yana
Unfortunately, in spite of Yana and MA1 yielding somewhat different admixture ratios, both ancestries are used indiscriminately as if they were mutually exchangeable, without checking which of the two yields better fits (e.g. Table S6.16 just gives p-values for the MA1 admix, while Fig. S6.9, S6.11 only include Yana).
As you state: “They show in different stats and figures in the paper that the Malta1 (near Lake Baikal, ca. 24 kya) shares more drift with Native Americans than the Yana samples. And AfontovaGora3 (Altai region, 17 kya) shares even more drift with Native Americans than Malta1 does.” I suppose you refer to Tab. S6.5 & S6.8 here, which indeed display said result.
OTOH, when looking at their qpWave/qpAdm results in Tab. S6.13, Yana outperforms MA1 and AG3 as secondary source in the Surui aside from Han. All attempts to model Surui as a two-way admixture from Tianyuan and a North Eurasian source fail to produce sufficient rank, meaning that at least one more admixture component is required (a result that mirrors the finds of Skoglund e.a. 2015). Nevertheless, both Yana and MA1 produce already reasonably good fits in the Tianyuan-Surui exercise, while AG3 is clearly rejected. Strange, isn’t it? D-/f-stats favour ANE, qpWave/qpAdm favour ANS.
One possible factor that comes to mind is Yana apparently having received extra Neandertal admixture compared to MA1 (c.f. Fig. S6.3). I initially thought “of course, Neandertal admixture decays over time, hence the difference“. However, Fig S6.8 (SI p. 97 – their numbering is inconsistent, they have two Fig. S6.8) shows high Neandertal admixture also in AP and Paleosibirians, several UstBelaya_EBA samples even exceed Yana in Neandertal affinity. Apparently, time decay of Neandertal ancestry isn’t much of an issue in NE Asia. Instead, we might be dealing with some Neandertal-specific genetic adaptation to arctic climate, which was evolutionary advantageous in the region, while eradicated in more moderate climates.
Fig. S6.9 (SI p. 98) shows:
1. An (expectable) decay of the average track length of archaic ancestry segments the younger the samples: UI nearly 350 kb, Sunghir/ Yana 200-240 kb, Anzick_1/ Bichon/ Devils Cave ca. 155 kb, modern Amerindians / Iran_N ca. 140 kb (the latter possibly reflecting earlier Neandertal admixture in the Near East than in Siberia), modern French ca. 100 kb.
2. Considerable regional variation in the share of archaic ancestry – Yana/ AP 2.2-2.4%, Amerindians (ancient & modern) 1.7-2.0%, CHG/ANF/EEF/ French 0.7-1.2%.
Unfortunately, neither MA1 nor AG3 are included, and they have used a specific calculation (or filtering?) method that yields quite different results to those provided in Fu e.a. 2016 (e.g. Stuttgart with them has 1.1% archaic ancestry, compared to 2.1% in Fu e.a., UI 2.7% vs. 3.0%, Karitiana 1.8% vs. 2.1%). That leaves wide room for guessing where MA1 / AG3 would plot. It clearly should fall closer to ancient and modern Amerindians than Yana, but how much closer, and whether that alone can explain that ANE is preferred over ANS in D- and f-stats is impossible to say.
I found their paleo-climatic simulation very interesting, and think it provides good background, also to your questions, Rob. Apparently around 38 kya, their estimated date of the ANS split from Early West Eurasians (EWE), the best place to be north of Lake Baikal was the Ob basin, right up to its estuary. Upper and middle Yenissei and Lena were also o.k., as long as you didn’t move too far north. By 32 kya, conditions had deteriorated in the Ob basin, while now the Lower Yana basin enjoyed very favourable climate conditions, better than even on the middle and lower Amur. Over the same period, Manchuria, i.e. the area north of Tianyuan, and the Sea of Okhotsk coast turned from “quite o.k.” into “hardly inhabitable”. That should have led original EWE settlers on the Ob to move eastwards with the shifting climate to the Upper Yana, where they encountered East Asian climate refugees to jointly form ANS. When assuming that population density mirrors climatic favourability, the Yana samples are not just the result of a tentative colonization attempt, but should be fairly representative of ANS.
6 kya later, the East Siberian climate had somewhat leveled. The Lower Yana, while still being among the better places to be, had lost a lot of its favourability. Conversely, conditions had improved again in Manchuria and around the Sea of Okhotsk, and Beringia started to become inhabitable. This should have resulted in ANS dispersing into all directions, including, but not limited to, the Baikal Lake region.
22 kya, at the onset of the LGM, things start to get really tough on the Lower Ob. The Lena basin and the Lower Yana benefit somewhat from the emerging Beringian microclimate. However, the biggest “winners” lie further east, both along the modern northern and southern Siberian coastline, and around today’s St. Lawrence Island south of the Bering Strait. The Kolyma river replaces the Yana river as climatically most favoured connection between Arctic Ocean and the Sea of Okhotsk. Animal herds, and HG following them, should have moved with the climate, and that means rather (south-)east than southwestwards.
For those who didn’t move, it was soon all over. When the LGM set in, things got pretty bad on the north coast and only turned better again around 14 kya, before the Lower Dryas delivered the next blow. The Chatanga-Kotui basin and the Upper Lena basin provided some initial refuge, but where you really wanted to be during the LGM was either East Beringia, or the wider Lake Baikal area (in fact, rather a bit further south, in the Upper Amur catchment area, or a bit west in the N. Altai). Things got equally bad between Upper Lena Basin and East Beringia, and the latter became virtually isolated.
Finally, when things gradually got better in Central Siberia and further west from 18 kya on, the Beringian microclimate disappeared, and with it the Beringian refugium that was all gone 14 kya latest. There was hardly a way back – Kamchatka and the Sea of Okhotsk were equally bad off. Wrangel Island and NW of it fared a bit better (and had mammoths!), but the best way to go was clearly SE along the American coast.
That whole story makes a lot of sense to me and aligns well with the aDNA as well as with their demographic analysis that I will cite in the following from p 8f of the paper (that’s the part you missed there):
– “We find (..) divergence of Kolyma1 from the Ancient Beringian / Native American ancestral population at ~24 kya.”, i.e. around when the Lower Yana loses its climatic advantage and starts to be overtaken by Beringia
– “ “Both Kolyma1 and Native American ancestors received ANS-related gene flow at a similar time (Kolyma1 20.2 kya (15.5-23.7); USR1 19.7 kya (13.3-23.5))”, i.e. around the peak of the LGM, when surviving ANS finally had to retreat to either the Upper Lena basin or Beringia.
If there weren’t the a/m D-/f-stats, I had little doubt about Yana having been the Ancient Native American “basecamp”. Genetic isolation of Ancient Beringians/ Native Americans after ca. 24kya has already been demonstrated in various other studies, so MA1 (and certainly AG3) is too late to be directly ancestral.
I question their assessment when it comes to AP, though. Having more or less the same amount of archaic admixture as Yana, the “more Neandertal” argument from above can’t explain AP’s preference for Malta over Yana. Moreover, in Fig. S6.8 (SI p.84), EHG (Karelia) gets an even higher f3 score than Malta, which is suggestive of some CHG admixture in AP.
In fact, there is an obvious CHG connection to East Siberia and beyond, in the form of mtDNA X2. X2, as its “nieces” U and HV, descends from “West Eurasian” mtDNA N, and has its highest Eurasian frequencies among Druzes and Georgians. X2 is earliest reported from Zagros_N (early 8th mBC), to afterwards regularly occur in AAF, EEF, CA/ BA (Armenia, Jordan, Remedello, Iberia) and BB.
X2a, however, has so far been exclusively found in N. America. While attestation in Kennewick Man (ca. 7k BC) demonstrates early presence in the Americas, X2a is unlikely to have arrived with the initial Beringian expansion. Soares e.a. 2009 (https://www.cell.com/ajhg/fulltext/S0002-9297(09)00163-3) estimated the age of X2a at 12.8 kya, substantially younger than the now generally accepted age of 14.8 kya for the Paleo-American settlement in Monte Verde, SC. Chile. The fact that X2a has so far only been found in N. America (the southernmost modern attestations are from the Central Mexican Pacific coast) also speaks against having participated in the initial expansion. [Intriguingly, the highest X2a frequencies, up to 25%, have been reported for Algic speakers, whose homeland is assumed to have been around Kennewick]. Last but not least, in the PCA of the ANS study discussed here (displayed in the original post), Kennewick Man plots more or less half-way in-between Neo-Siberians (Ekven_IA, Uelen_IA) and Native Americans, with a slight but well visible pull off the East Asian – Amerindian cline towards Zagros_N.
In short – I think AP may well contain some CHG admix.
That takes me to my final point – the asymmetrical relation of Mixe and Pima to Yana, suggestive of some later genetic shift (great observation, Alberto, btw, I would have overlooked it). Now, there is ample indication of Chinese contact with Pacific Mexicans, including a 5th cCE written Chinese source, the Chinese Crested Dog sharing a rare gene mutation with the Nahuatl (Aztec) Xoloitzquintl and the Peruvian Hairless Dog, and NW American wolf DNA attested in several traditional Chinese dog breeds. But, of course, such contact doesn’t show up if you try to model Native American Mexicans as admixture of ANS/ ANE and Han (or Tianyuan, for that matter), as the study discussed here did.
What, however, shows up in Tab. S6.13 is that both ANS and Malta fail as source populations when attempting to model Mixe in qpAdm as a two-source admixture with Han as second source. OTOH, EHG_Karelia does the job, with a pretty good model probability. So, here we go again with additional CHG input!
EHG_Karelia is modeled as 24% WHG, 76% pre-EHG (WSHG?), which in turn is modeled as 78% ANE, 22% CHG, yielding 16.7% CHG in EHG_Karelia. Their qpAdm run has Mixe as 25% EHG_Karelia, which ultimately means some 4.2% CHG on top of whatever mix of Paleoindian (EEA – ANS) and IA Chinese ancestry Mixe hold.
If true – from where and when did that CHG admix arrive? One possibility is “trickling south” of the CHG component that I suppose to be manifested in Kennewick Man / Algic mtDNA X2a. But in that case, shouldn’t there be at least one or two attestations of X2a in Mixe or Pima? Well, I couldn’t find any in the available literature. Moreover, why should “trickling south” of such ancient admixture apparently have stopped in Mexico and not reached further south, while we have multiple evidence of genetic exchange between Mexico and South America since the mid-Holocene as concerns crops such as maize, cotton and chili peppers?
Even more intriguing is that EHG_Karelia as well-fitting admixture source into Mixe contains 24% WHG, translating into 6% WHG admixture in Mixe. So, we seem to ultimately be dealing with an admixture source into Mixe that was approx. 60% WHG, 40% CHG. Such a source looks fairly West European/ North African, but I don’t have any idea what that source might have been. I briefly thought about Punics, but they should have been less WHG and much more Natufian (Basal Eurasian). In conclusion, while their results point at a trans-atlantic admixture source into Mixe, further modeling and analysis will be required to sort out details.
Alberto: Whether this is because the split from West Eurasians soon after the split between WE and EE alone
In theory, a basal split from other WEu would just mean an equivalent of how Tianyuan is to the ENA clade; basal to all other lineages (and then admixing with a population that clades deeply with Onge to give rise to East Asians, as in the model in McColl 2018 – https://imgur.com/a/O2TmsPb). That wouldn’t make D(Mbuti,Tianyuan;ANS,K-14) or D(Mbuti,Onge;ANS,K-14) different from 0. C’mon, this is stuff you know :).
It’s got to be a “complex split” between early branching clades, meaning that the early split of East and West Eurasians was probably characterised by multiple lineages around 50-40 kya that split and merged in a cloud that is hard to recapitulate, until the main West-East split emerged, along with some stats that are hard to explain among them (e.g. GoyetQ116-Tianyuan attraction).
Your point around accelerating f4 drift Yana->MA-1->AG-3 with Native Americans and how that probably indicates a lack of an isolated ANS source contributing to NA is well made and reasoned, btw. I don’t know if this necessitates extinction as such, but certainly it seems to preclude total continuity from the time of these samples as *the* ANE source for NA.
These Yana samples seem pretty basally divergent within the clade that leads to ANE, shared f3 (page 64) is quite high with Yana to MA-1, but when it comes to AG3, AG3 is quite a bit down the list compared to other European Upper Paleolithic populations (though I would note weak SNP overlap). AG-3, on the other hand has a strong relationship with MA-1 in the paired f4 stat, f4(Mbuti,X;MA-1,Yana).
It looks like Yana has a strong relationship to MA-1 and MA-1 to AG-3, but AG-3 does not have such particularly strong relationship to Yana.
The qpAdm / qpWave results where Yana seems equally or more viable as a contributor to Native Americans look to me like a lack of discrimination in the outgroups.
It would’ve been interesting to see the West_Siberia_N and Botai samples tested as well. Okunevo shows a weak relationship to Yana samples in the fig on p65, but both the West Siberia N and Botai should be quite significantly more ANE and quite low of East Asian related.
@Matt, nice to see you!
Yes, you are right that a split between ANS and WEu soon after the split between WEu and EEu wouldn’t explain the higher affinity to EEu in ANS (which is probably why I added: “though in theory I guess that admixture is the only way to explain a higher affinity to EE (or otherwise WE admixed into some other more divergent ghost population that separated them further from EE, but this is quite more speculative).” But I disagree that this is stuff that I know (half of the times that I write about these complicated things I have to wonder if I’m saying something stupid, so it’s good to get confirmation from others).
It’s probably a more complex split scheme as you suggest, with some admixture between different groups that makes it difficult to know very precisely how it happened.
Re: extinction, I think this is something that happens frequently both with animals and humans. When the climate changes and makes it hard to survive in a certain niche, it’s not that easy to migrate to find a better one (most of the times, especially talking about the area in question, that would be a suicide). I guess that survival is more about being lucky enough to be in the right place, rather than smart enough to move to the right place in advance by predicting the upcoming changes (both for animals and humans). But yes, ultimately we can’t say with certainty if any of those groups did survive and later merged with other incoming populations, since they were related enough to be too difficult to detect such admixture in the later groups.
@FrankN
Yes, I agree that Ancient Paleosiberians and Native Americans are part of the same process, and not the same population (or more specifically, Native Americans were an early subpopulation of Ancient Paleosiberians that migrated to the Americas, with later groups in Siberia being more East Asian shifted). I don’t know if I implied that somewhere in the post, but if I did then your correction is… correct.
I (probably, will have to check them again) agree with what Matt said above about the qpAdm models not having enough discriminatory power with the outgroups, because otherwise the D-stats look quite clear about MA1 and AG3 being better sources for NAs.
I’d also like to check in detail the rest of the things you comment about (the mtDNA X2, EHG/CHG, etc…) That’s an interesting thing to look at, so I’ll do as soon as I can to be able to comment further.
Alberto, Frank & Rob,
I have raised this issue elsewhere but I am going to expand a bit upon it.
I am wondering about the linkages between AASI, Iran_N/CHG and ANE ancestry. I am wondering whether there is a considerable amount of shared ancestry between the 3 ancestry components.
I have wondered whether Iran_N has some AASI ever since having seen the admixture graph of the following Gallego Llorente paper –
https://www.nature.com/articles/srep31326/figures/1
As can be seen here, GD13a primarily seems to consist of two admixture components, a major one which it shares with CHG and a minor one which maximises in Indian tribal and South Indian groups and which therefore seems to closely parallel the AASI.
I also take the liberty to post Alberto’s comments around that time on Davidski’s blog,
Another thing I wanted to check was if Iran_Neolithic has ASI. It’s not easy to check directly with the populations available, but starting from a model for Kalash (no Onge, so using Paniya):
Kalash:HGDP00267
“Iran_Neolithic:I1945” 47.2
“Yamnaya_Samara:I0231” 38
“Paniya:PNYD1” 14
“EHG_Karelia_HG:I0061” 0.8
“Anatolia_Neolithic:I0707” 0
“Israel_Natufian:I1072” 0
“Mal’ta_AfontovaGora3:I9050.damage” 0
“Mal’ta_MA1:MA1” 0
“Levant_Neolithic:I1699” 0
“Villabruna_Loschbour:Loschbour” 0
“Yoruba:HGDP00920” 0
“Ami:NA13607” 0
“Satsurblia_Kotias:KK1” 0
“Andronovo:RISE505” 0
“Dai:HGDP01307” 0
distance=0.003709
Paniya is quite low compared to what we’re used to see. And then removing Paniya:
Kalash:HGDP00267
“Iran_Neolithic:I1945” 62.45
“Yamnaya_Samara:I0231” 15.9
“Mal’ta_MA1:MA1” 14.05
“Villabruna_Loschbour:Loschbour” 6.2
“Dai:HGDP01307” 0.9
“EHG_Karelia_HG:I0061” 0.5
“Anatolia_Neolithic:I0707” 0
“Israel_Natufian:I1072” 0
“Mal’ta_AfontovaGora3:I9050.damage” 0
“Levant_Neolithic:I1699” 0
“Yoruba:HGDP00920” 0
“Ami:NA13607” 0
“Satsurblia_Kotias:KK1” 0
“Andronovo:RISE505” 0
distance=0.006984
Iran_Neolithic roughly takes the Paniya, and there is hardly any ENA in the model, just traces of Dai. The model is worse, but still quite good.
Trying to model Paniya:
Paniya:PNYD1
“Iran_Neolithic:I1945” 69.15
“Mal’ta_AfontovaGora3:I9050.damage” 16.85
“Dai:HGDP01307” 14
…
distance=0.043548
A poor model, but still surprising. It seems like Iran_Neolithic does have some good amount of ASI. We’d need to check this with some stats.
As is evident, this above analysis by Alberto also seems to support AASI in Iran_N.
With time, I have begun to think that this AASI ancestral component in Iran_N is not likely due to some pure AASI population admixing into Iran_N. Rather it appears to me that this could be a sign of some deep ancestral links between Iran_N & AASI.
There was this recent aDNA paper on late Bronze age & Iron age samples from the steppe
http://advances.sciencemag.org/content/4/10/eaat4457
Figures 2C & 3D show the presence of a South Asian ‘lilac’ component at k=15 that is maximised in South Indian tribal groups (figure S10). Here, in this paper, this ‘AASI’ component is also found in CHG and various steppe groups.
However, this admixture graph ‘evidence’ has to be supported by other lines of evidence such as the f-stats which so far, due to lack of appropriate samples, was not forthcoming.
———————————
Now recently there were also two important papers on Palaeolithic DNA – one from Anatolia and the other one from Dzudzuana.
In the Anatolian Hunter Gatherer paper the following is of interest –
The authors model AAF (Anatolian Aceramic Farmer) as 90 % AHG + 10 % Iran_N.
In the paper it is stated at one point (pg 5, last para) –
In turn, AAF is slightly shifted upwards compared to AHG in the PCA, to the direction where ancient and modern Caucasus and Iranian groups are located. Likewise, when compared to AHG by D(AAF, AHG; test, Mbuti), the AAF early farmers show extra affinity with early Holocene populations from Iran or Caucasus and with present-day South Asians, who have also been genetically linked with Iranian/Caucasus ancestry14, 15 (Fig. 2A, fig. S2 and data table S3).
Since, I did not have access to the Supplementary Data, I will only refer to figure 2A on pg 24. In the inset, one can see the top results of the stat D(AAF, AHG; test, Mbuti).One can see that while Kotias is the topmost, it is followed by Vishwabrahmin and Mala, who are heavily AASI and have only a minority of Iran_N. Iran_N itself comes further down the list.
So this stat seems to suggest that whatever population admixed into AHG to form the AAF (Anatolian Aceramic Farmer), also substantially increased its affinity towards heavily AASI groups as well as Iran_N & CHG. This supports the argument that Iran_N, CHG & AASI may have some deep ancestral linkages and which is also being reflected in the admixture graphs at higher Ks.
Moving on to the data from the Dzudzuana paper –
In the table 1 (pg 13) of the Dzudzuana paper, one can see the best qpAdm models for various ancient samples.
For Iran_N, the qpAdm model is
Mbuti : Onge : AG3 : Duzzuana =
0.097 : 0.109 : 0.218 : 0.577
while for CHG it is
Mbuti : Tianyuan : AG3 : Dzudzuana =
0.054 : 0.081 : 0.222 : 0.643
So one can clearly see that CHG and Iran_N can be shown to have East Eurasian related (Onge/Tianyuan) related ancestry of 8.1 % & 10.9 % respectively. Frank has argued that the source of East Eurasian admixture in CHG and Iran_N is different. However that looks unlikely because the source is likely to neither be Onge or Tianyuan but more probably AASI since the East Eurasian Ancestry proportion in these two stats for Iran_N & CHG corresponds well with the small admixture component found in Iran_N and CHG in admixture graphs which I had alluded to earlier and which is maximised in South Indian tribals i.e. it corresponds with AASI.
Further, in table S3.2. pg 39 of the Supplementary Section of the same paper, we can see that Anatolian_N can be modelled as a mixture of
Onge/Tianyaun/Papuan/Han/Ust-Ishim + Dzudzuana
which supports what we found in the Anatolian Hunter Gatherer (AHG) paper where AAF or Anatolian_N showed greatest shift to Vishwabrahmin & Mala after Kotias relative to AHG.
Combining all the data it appears, in my humble opinion, that there seems to be a solid case for substantial levels of shared ancestry between Iran_N/CHG and AASI. The origin of this shared ancestry has to be sought to the east of the Near Eastern region, since besides Iran_N & CHG, none of the other ancient Near Eastern groups have this AASI-related component.
——————————————-
Having come late to the party, I just saw a couple of days ago that Alberto had posted the following in the Dzudzuana post,
CHG
Sarazm_Eneolithic 82.9%
Tepecik_Ciftlik_N 17.1%
Ganj_Dareh_N 0%
Barcin_N 0%
Boncuklu_N 0%
Kostenki14 0%
Vestonice16 0%
AfontovaGora3:I9050.damage 0%
EHG 0%
Comb_Ceramic 0%
Latvia_HG 0%
Baltic_HG 0%
Narva_Estonia 0%
Narva_Lithuania 0%
WHG 0%
Distance 13.9802%
The thing is that the North Caucasus steppe population and Yamnaya-CWC-BB have some mtDNA clades that are steppe markers for Europe, but we also find them in SC Asia (J1b1a1 in Geoksiur Eneolithic, w3a1 in Tepe Anau Eneolithic, T1a1 and W6 in Gonur Tepe BA), which suggests an ancient connection between CHG and SC Asia (West Iran has not shown this connection through mtDNA so far).
On a related note (probably for another future post), I was also wondering if there’s genetic evidence of migration from Neolithic West Iran to North India. This has been given for granted so far, but I’m not sure why. The genetic structure of the Indus_diaspora samples could be defined as Iran_N + ANE + AASI. If one proposes a migration of Iran_N, you’d need a local population that were pure ANE and pure AASI or a mix of both already. I don’t think we’ll find that in Mesolithic North India. The alternative is a model of something more closely related to Sarazm_Eneolithic + AASI:
Shahr_I_Sokhta_BA3
Onge 51.2%
Sarazm_Eneolithic 43.8%
Ganj_Dareh_N 5%
AfontovaGora3:I9050.damage 0%
EHG 0%
Naxi 0%
Distance 13.0729%
So is there shared ancestry between Iran_N and Indus_diapora or is there migration from Iran_N?
Here the close relationship observed between Sarazm_EN (which has small levels of AASI) and CHG further supports my point that the origins of CHG/Iran_N might be around SC Asia.
——————————————-
Now let us bring ANE into the picture.
In the Dzudzuana paper, as per table S3.2, AG3 can be modelled as a two way mixture of MA1 + Ust Ishim/Papuan/Onge/Han/Tianyuan
As per this current ANS paper, MA1 can be modelled as 72 % Yana + 28 % CHG, while EHG can be modelled as 19 % CHG + 81 % MA1.
Combining this with the earlier qpAdm models from the Dzudzuana paper where Iran_N & CHG both showed AG3 admixture along with the ENA admixture we observe that –
– Iran_N and CHG both seem to have AASI & ANE ancestry.
– The ANE also has some ENA ancestry (which could be AASI) and some CHG/Iran_N related ancestry.
It may also be noted in passing that Iran_N does have y-dna R2 which cements its links with ANE ancestry. The Iranian plateau and the Indian subcontinent also share some very deep y-dna Q lineages.
So this raises a very important question – Is there an trilateral intersection of ancestries between ANE, Iran_N/CHG and AASI ? In other words, do they have substantial levels of shared ancestries ? The answer to this question can be of vital importance. It may mean, among other things that the geographical origin of ANE, Iran_N/CHG & AASI ancestries may be quite similar.
Alberto:
“Native Americans were an early subpopulation of Ancient Paleosiberians that migrated to the Americas..”
That is still somewhere besides the point the paper, IMO correctly, makes. As per their demographic modelling (Fig. 3b), ca. 24 kya a yet unsampled Early Middle Upper Paleolithic East Asian (EA_EMUP) population, related to TianYuan, split into Ancestral Beringians (as a ghost inferred from the 9.5 kBC Upward Sun River sample from Alaska, and Anzick1) and a Later Middle Upper Paleolithic East Asian (EA_LMUP) population (as a ghost inferred from Kolyma1). That split date corresponds to earlier studies postulating a certain period of Beringian genetic isolation, before they moved onwards into the Americas.
After that split, both of EA_LMUP and Ancestral Beringians received admixture from ANS, turning EA_LMUP into AP, and Ancestral Beringians into Ancestral Native Americans. Admixture proportions were somewhat similar, but not identical, and as concerns admixture times, they remain rather ambiguous, to put it nicely. For the ANS admixture into EA_LMUP, their Figs 3a and 3b provide two dates, 25.8 and 20.2 kya, respectively – most likely, this was a longer-term process that took place across a relatively large area around the Kolyma, Lena and Yana river basins and the Sea of Okhotsk. ANS admicture into Ancestral Beringians is inferred for somewhat later, namely 19.7 kya. This corresponds to the peak of the LGM, and may plausibly relate to populations from the Arctic Sea coast seeking refuge in East Beringia.
As such, it is not one population (Ancient Native Americans) being a subpopulation of the other (AP). It is the same process, namely the onset of the LGM leading to two previously differentiated populations, ANS and EA_EMUP, seeking refuge in the same places and admixing there. But this process occured in different geographical areas, at a somewhat different time, with different admixture ratios, and also among populations that already had separated and genetically drifted away from their common ancestry (EA_LMUP – Ancient Beringian split).
ANE is so far defined by MA1 and AG3, which implies a timeframe of ca. 24-16 kya, and a Central Siberian location. The process described above commenced earlier, at least as concerns AP formation. Moreover, as per the study’s climatic modelling, there is only one feasible route how an Early West Eurasian-like population could have entered Beringia by the onset of the LGM, and that is the along the Arctic Sea coast, not from anywhere around Baikal Lake. As such, it is methodologically sound to assign a fresh designation to that source population, namely ANS. Whether the Lower Yana samples, the first samples available so far, fully capture the ANS genetic profile, or other, more ANE-like ones to be (hopefully) sampled in future may be more representative is another issue. Nevertheless, when having to choose between re-defining ANE towards a much wider chronological and geographical coverage, or introducing ANS as a new designation, even if at the moment defined by only two samples, my personal preference clearly goes towards the latter.
At the moment, the oldest mtDNA X in the ancient record is Iran_Neolithic Ganj Dareh I1290 X2, 8179-7613 calBCE (8780±50 BP). Kennewick man with X2a is estimated to have lived 8,358 ± 21 14C years bp. Therefore, they are almost equally old. X2a is defined by a HVR1 mutation at G16213A, therefore, it is relatively easy to detect.
Considering the distribution of mtDNA X, it may have arisen in (South) Central Asia.
@FrankN
Ok, I see your point now. It seems I overlooked that part about the split between EA_EMUP and EA_LMUP, which is why I didn’t understand well the statement about the “genetic isolation between Asians and ancestral Native Americans after ~24 kya” as I said in the post. I hope I can get around looking into this and all the other details you mentioned earlier.
Still, for the ANS-ANE part, it seems that if there was an ANS-related population closer to Beringia that gave rise to Native Americans, such population would form a clade with AG3 to the exclusion of ANS, and therefor it would still be more apt to call that putative population ANE rather than ANS (and ANS in this case would just be an isolated population that didn’t contribute to later populations). But this scenario of a population near Beringia being closer to AG3 than to ANS seems a bit of a stretch (especially when we have MA1 in between -both geographically and temporarily- that also shares less alleles with Native Americans than AG3 does.
@Jaydeep
You’re raising some complicated questions that are ultimately quite difficult to answer (and i don’t even have the right tools to test possible scenarios about it).
I’d say that an ancient relationship between CHG, Iran_N and Sarazm_En seems very likely. Also all of these populations have a high amount of ANE admixture, and therefor an ancient connection to ANE.
The most difficult part is to know if AASI has any connection to either of these populations. And this is difficult to answer until we get relevant samples.
A possible scenario would be that AASI and ANE formed in geographic neighbourhood and influenced each other. Given the time frame we might be talking about (40-35 kya), somewhere closer to South Asia seems more plausible than in more northern latitudes. I can imagine that AASI might turn out to be not something 100% ENA, but just like now we’ve come to know that ANE is not 100% WEu, AASI could turn out to be the opposite in those terms (~75% ENA and ~25% WEu). But for now this is just a speculative scenario like any other. Besides, we’re talking about some very early connections, so even then it’s hard to say how they might be influencing more modern samples.
I can only say that we have to wait and see how all of this turns out as we get relevant samples (which I don’t expect to be really soon, given the area and time we’re speculating about).
@ Alberto
There are some very old remains (c. 30 ky BP) from Sri Lanka.
It would not be too much of a stretch to imagine that attempts to extract aDNA are being undertaken.
“This suggests that whatever Australasian “admixture” (if it can be called admixture at all) present in modern samples from the Amazon might have been already present in the Anzick-1 sample, and it’s rather Mixe and Pima who would have later experienced some genetic shift that makes them further away from Australasians (but apparently IIRC, there’s no evidence of admixture from any known source in Pima and Mixe, which makes the case complicated”
That’s the suggestion in various non-formal analyses I’ve seen with Pima and Mixe having some sort of Paleosiberian ancestry that Karitiana lack but that might be an artifact. You might be right though I think there’s a forthcoming paper that will argue the signal in South America is real.
@Jay, I think you’re right that ADMIXTURE tends to see a small AASI like (centered in South Asia but shared with Southeast Asia to an extent) component in Iran_N with the rest being a unique component shared with CHG, with the latter having some ANE/EHG kind of stuff on top of it. Hard to know if it stands for something shared or arriving from one area to the other but interesting speculation.
I’d say that an ancient relationship between CHG, Iran_N and Sarazm_En seems very likely. Also all of these populations have a high amount of ANE admixture, and therefor an ancient connection to ANE.
The most difficult part is to know if AASI has any connection to either of these populations. And this is difficult to answer until we get relevant samples.
In my humble opinion, the relationship with AASI seems very likely and it is not based on heavy speculation.
In the Dzudzuana paper, it is shown that Anatolian_N can be modeled as Dzudzuana ( 90 + %) and either of Ust-Ishim, Onge, Han, Tianyuan. i.e. in comparison to Dzudzuana, there is clearly an East Eurasian shift in Anatolian Neolithic.
Coming to the AHG paper,
Anatolian_N or AAF are modelled as 90 % AHG & 10 % Iran_N. Now AHG is likely to be the closest ancient population to Dzudzuana for a variety of reasons. So if we consider AHG & Dzudzuana as identical, it may be argued that an Iran_N like admixture in AHG shifted to towards East Eurasian ancestry. That Iran_N and CHG have East Eurasian admixture is confirmed by the 4 source model of Iran_N & CHG in the Dzudzuana paper.
So it looks quite certain that Iran_N and CHG brought East Eurasian admixture in the Near East and also spread it into Anatolia_N.
Now the question is which East Eurasian source could this be ? We may note the following :-
When one looks at Iran_N and CHG in the admixture graph of the recent steppe paper which included Srubnaya Alakulskaya samples, a component that maximises in South Indian and Tribal groups is also present at about 10 % or so in both of them. This is quite different from other East Eurasian related ancestry components that maximises in East Asians, Onge or SE Asians and is clearly a component related to high AASI proportion groups.
So, could the East Eurasian admixture in Iran_N & CHG as per qpAdm in the Dzudzuana paper not be this very same AASI component observed in admixture graph of the Srubnaya paper ?
As a supporting proof, note that Anatolian Aceramic Farmers (AAF) or Anatolia_N when compared to Anatolian Hunter Gatherers (AHG) share most alleles with CHG, AASI heavy groups like Vishwabrahmin & Mala, Iran_N and then Pathan, Kalash etc.
So here again, the East Eurasian admixture observed in Anatolia_N by the Dzudzuana paper, and which most likely was mediated into Anatolia_N by an Iran_N like group, increases Anatolia_N affinity to AASI heavy groups and not some East Asian or Siberian group.
So the simplest explanation of this is that the East Eurasian admixture observed in Iran_N, CHG and Anatolia_N is AASI related.
And it looks like it was brought to the Near East by Iran_N/CHG like populations which may mean they have a more eastern origin.
A possible scenario would be that AASI and ANE formed in geographic neighbourhood and influenced each other. Given the time frame we might be talking about (40-35 kya), somewhere closer to South Asia seems more plausible than in more northern latitudes. I can imagine that AASI might turn out to be not something 100% ENA, but just like now we’ve come to know that ANE is not 100% WEu, AASI could turn out to be the opposite in those terms (~75% ENA and ~25% WEu).
Mostly agree but I may add here that AASI and ANE rather than influencing each other may just have significant levels of shared ancestry that diverged with the passage of time. We may note that ANE itself has East Eurasian admixture though at present we cannot identify the most likely source for it.
“This suggests that whatever Australasian “admixture” (if it can be called admixture at all) present in modern samples from the Amazon might have been already present in the Anzick-1 sample …”
But note Skoglund e.a. 2015 (full text available at https://reich.hms.harvard.edu/publications): “We do not detect any excess affinity to Australasians in the ,12,600-year-old Clovis-associated Anzick individual from western Montana (Z 5 20.6)”
Aside from the Australasian signal detected by Skoglund e.a., such admixture is also indicated by Denisovan ancestry in NE South Americans detected by Qin/Stoneking 2015 (https://doi.org/10.1093/molbev/msv141). In this context, I noted in the Sikora e.a. SI (p. 96) :”We find that all [newly sampled] individuals show the signal of Neanderthal admixture typical of non-African populations, without any noticeable affinity to Denisovans beyond that due to its shared ancestry with Neanderthals “, which means that the Denisovan ancestry most likely didn’t reach the Americas from NE Siberia (including Devil’s Cave). The main text furthermore notes that no Australasian signal was found in any of the fresh samples (and also not in the Alaskan USR sample). As such, a scenario of a sub-structured Ancient Beringian / Native American population that included ancient Australasians, as considered in Skoglund/ Reich 2016, is hardly tenable anymore.
While Polynesian contact with S. America is evidenced, Skoglund e.a. 2015 ruled it out as (primary) source of the Australasian signal – their linkage disequilibrium analysis pointed at the admixture being substantially older than 4kya. Roewer e.a. 2013 (https://doi.org/10.1371/journal.pgen.1003460) identified a distinct geographical cluster of yDNA C-M217 (C3*) on the Pacific coast of Ecuador and Columbia. Otherwise unreported from the Americas, but frequent a/o among Koryak and Hokkaido Ainu, and at an estimated admixture time some 6kya, they putatively connected C3* to a Jomon culture contact with 4th mBC Ecuador that has been proposed by some archeologists because of apparent parallels between Jomon pottery and that of the Ecuadorian Valdivia culture. That proposal may hold true or not – we will sooner or later get aDNA from both cultures – but the C3* looks rather “Neosiberian” to me, and as such unlikely to have conveyed Australasian/ Denisovan ancestry.
A different issue is the Polynesian mtDNA that Faria Gonçalves e.a. (https://doi.org/10.1073/pnas.1217905110) have recovered from remains of Brazilian Botocudo Amerindians who, before their extinction in the late 19th century CE, lived not too far away from Surui and Karitiana. However, as the authors themselves warn, admixture time is uncertain, and there is the possibility that the Polynesian mtDNA was introduced by 19th century slaves of Malagassy origin.
Certainly a good paper. It poses some questions, such as when Ust’-Ishim’s population was replaced by a Yana-like population in western Siberia, the possibility of an admixture related to the former in the latter (although unlikely), the location in which Yana’s Western Eurasian and ENA ancestors met, the nature of the CHG-like component in ANE (which could be Dzudzuana, although I have not been able to test that because I don’t know how to use github files, leaving me unable to use qpAdm, and I’m still quite amateur), and other questions. The similarity between Yana, Mal’ta and Afontova Gora (both samples) suggests that ANE originated largely out of an ANS population likely living in southern Siberia. Another interesting thing is how similar the Dzudzuana paper’s modeling of ANE and this paper’s modeling of Yana are, with between 70% and 80% of a Western Eurasian component related to Kostenki and Sungir, and between 20% and 30% of an ENA component. I’m aware that this comment is partly speculation.
@Jaydeep
You’re pointing out many interesting things that might all add to prove your point. But I guess that until we don’t get more direct evidence it’s still uncertain.
Let’s see if relevant aDNA comes soon-ish and we can figure out all the details about AASI, ANE, CHG/Iran_N, etc…
I found your post and will read it as soon as I have a bit of time and comment there if I have anything to say:
http://www.brownpundits.com/2018/10/17/a-tentative-out-of-india-model-to-explain-the-origin-spread-of-indo-european-languages/
@FrankN
I had some time (though not too much) to look at those details you point out from the paper. Similarly to what Jaydeep wrote about a completely different subject, I think they’re all valid and further data (or analysis of the current data) might show they’re correct with more direct evidence. Apparently a paper is coming about the Australasian admixture in Karitiana and Surui that will clarify that part.
Let’s see if a closer look by someone who has the means at Kennewick Man and other lines of evidence (mtDNA X2a, possible admixture in Mixe and/or Pima, etc…) can tell us more about the history of Native Americans.
@Jaydeep:
Iran_N and AASI could have gradually diverged from each other during the HG stage with Iran_N being an earlier agriculture debutante vs the other.
A test would be to withhold Iran_N as admixture source and see if AASI rises
” I think you’re right that ADMIXTURE tends to see a small AASI like (centered in South Asia but shared with Southeast Asia to an extent)” — @ Egg, the AASI like component that’s seen in south-east asians in ADMIXTURE might be due to recent migration( in the last 2,000 years) from the indian subcontinent to south-east asia.